stringApp: stringApp: Importing and augmenting Cytoscape networks from string-db
stringApp imports data from string-db into Cytoscape. Users provide a list of one or more gene or protein identifiers, the species, and a confidence score and stringApp will query string-db and return the matching network. stringApp also allows users to expand the resulting network by adding an arbitrary number of nodes, change the confidence score, and expand the network by adding new terms. All resulting nodes are visualized using a new "String Style" custom graphic.
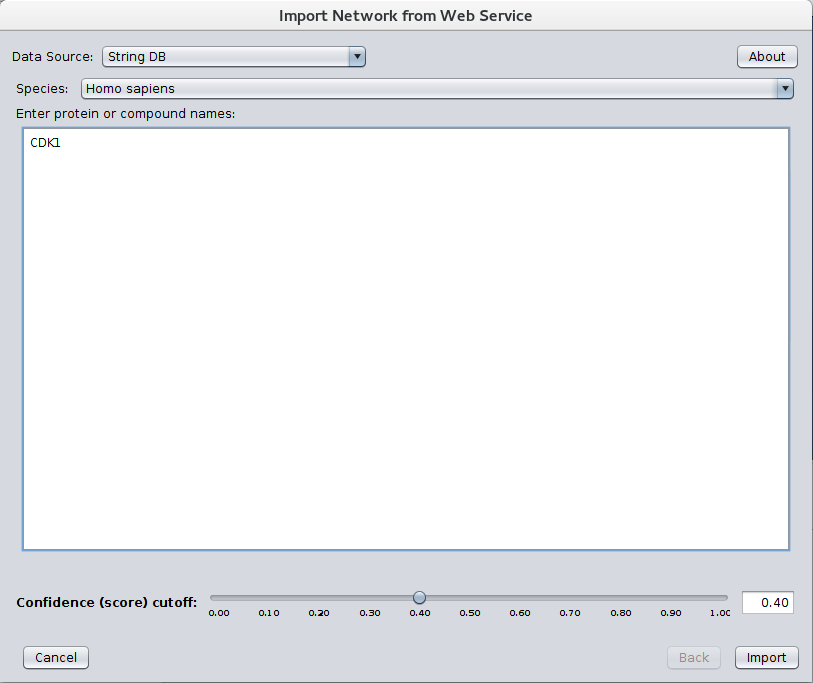
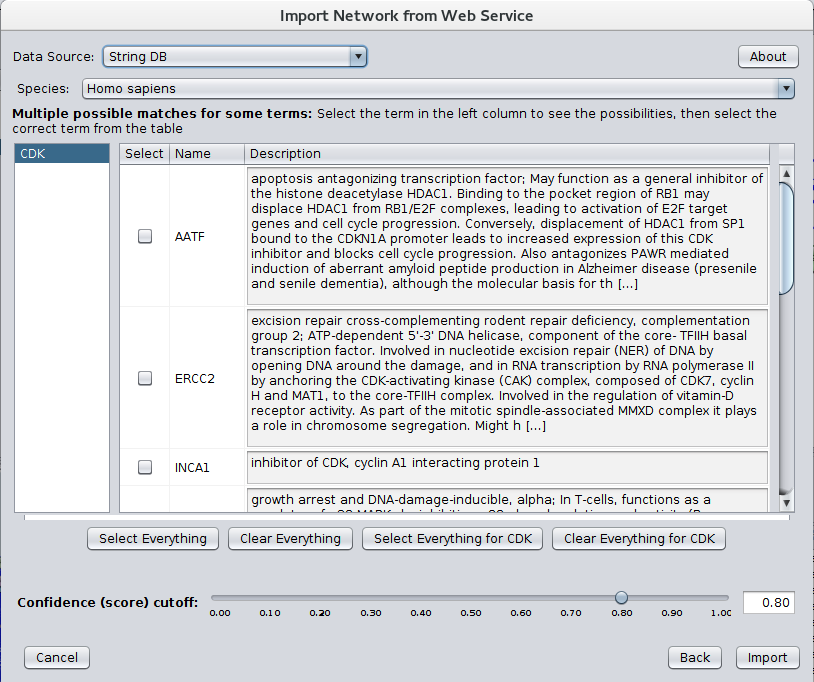
After installing stringApp from the Cytoscape app store STRING networks may be imported by going to File→Import→Network→Public databases... and selecting String DB for the Data Source. The dialog shown in Figure 1 should then appear. Select your Species and Confidence (score) cutoff and enter the genes or proteins of interest in the Enter protein or compound names text field. If multiple terms match any of the protein or compound names entered, a Term Resolution panel is shown (see Figure 2). This panel allows the user to select one or more terms (proteins or compounds) that match the query. Those terms will then be used for the query to STRING. If there is only one matching term, the term resolution dialog will not be shown and the matching term will be used directly. Sometimes one term may be a unique match while another term might require resolution.
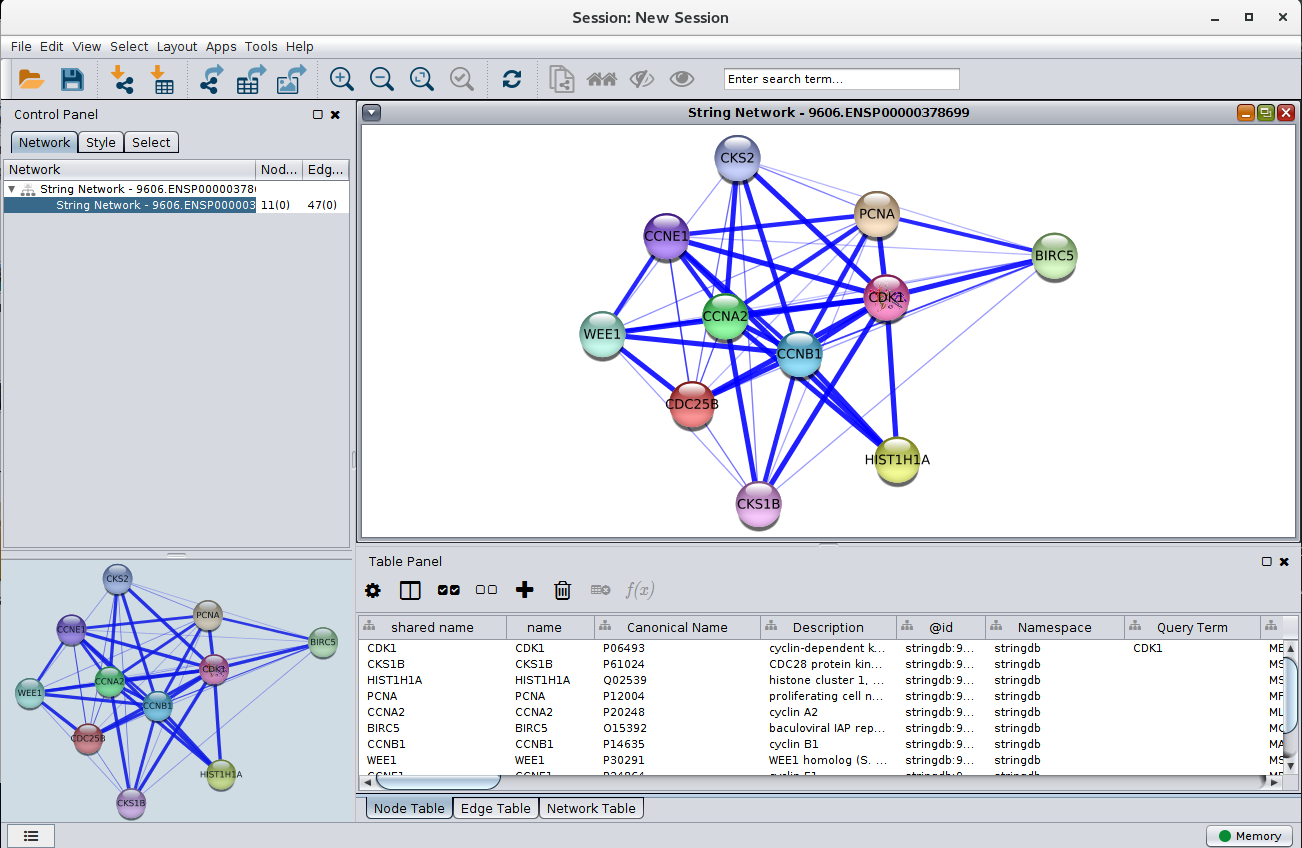
Figure 3. A STRING network loaded into Cytoscape. The query term for this network was CDK1, the species was Homo sapiens, and the score cutoff was 0.4.
Once the terms selected and sent to STRING, the resulting network is downloaded and visualized in Cytoscape. The node style is based on STRING's "glass" look, and maps to that look as closely as possible. If the STRING database includes any images for that node, they are downloaded also and included in visualization of the node. (NOTE: at this point, very few nodes include structural images. We hope to correct this as soon as possible). The widths and transparency of the edges are based on the aggregate score. All of the relevant scores are downloaded for each edge, including scores for: databases, text mining, experiments, coexpression, cooccurrence, and fusion.
Once the network has been imported and visualized, stringApp provides three ways to augment or adjust the network. First, the network may be expanded by adding additional nodes that match the confidence cutoffs. To add nodes, select Apps→String→Expand network and select the number of additional nodes to add. Select the Relayout network option to execute a force directed layout of the network after the new nodes are added. Note that any additional terms will be white when they are visualized, but will still retain the STRING styling.
The second option for changing the network is to change the miminum confidence score of the edges. Note that this does not fetch any additional data from STRING -- it only hides edges that fall beneath the score threashhold.
The final option is to add additional terms (proteins or compounds) to the network. This will bring up a network identical to the panel used to initially import from STRING. If the terms entered require resolution, the Term Resolution panel will be shown. Once the terms are selected and resolved (if needed), the nodes will be fetched and the user will be given the option to relayout the network. As before, nodes representing new terms will be colored white.
Publication
Cytoscape stringApp: Network analysis and visualization of proteomics data. Doncheva NT, Morris J, Gorodkin J, Jensen LJ. J Proteome Res. 2018 Nov 19. doi: 10.1021/acs.jproteome.8b00702. [PMID:30450911]
Last updated on January 23, 2019
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2021 Regents of the University of California. All rights reserved.