

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
Molecular insights into capsular polysaccharide secretion. Kuklewicz J, Zimmer J. Nature. 2024 Apr 25;628(8009):901–909.
Mechanical activation opens a lipid-lined pore in OSCA ion channels. Han Y, Zhou Z et al. Nature. 2024 Apr 25;628(8009):910–918.
Bitter taste receptor activation by cholesterol and an intracellular tastant. Kim Y, Gumpper RH et al. Nature. 2024 Apr 18;628(8008):664–671.
Structure and assembly of a bacterial gasdermin pore. Johnson AG, Mayer ML et al. Nature. 2024 Apr 18;628(8008):657–663.
Structural basis of DNA crossover capture by Escherichia coli DNA gyrase. Vayssières M, Marechal N et al. Science. 2024 Apr 12;384(6692):227-232.
More citations...News
January 22, 2024
ChimeraX 1.7.1 is available, with fixes for a few miscellaneous bugs that were identified after the 1.7 release.
December 19, 2023
The ChimeraX 1.7 production release is available! See the change log for what's new. Future Mac releases will require macOS 11 or higher.
November 6, 2023
The ChimeraX 1.7 release candidate is available – please try it and report any issues. See the change log for what's new.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325, Chan Zuckerberg Initiative grant EOSS4-0000000439, and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Feature Highlight
|
|
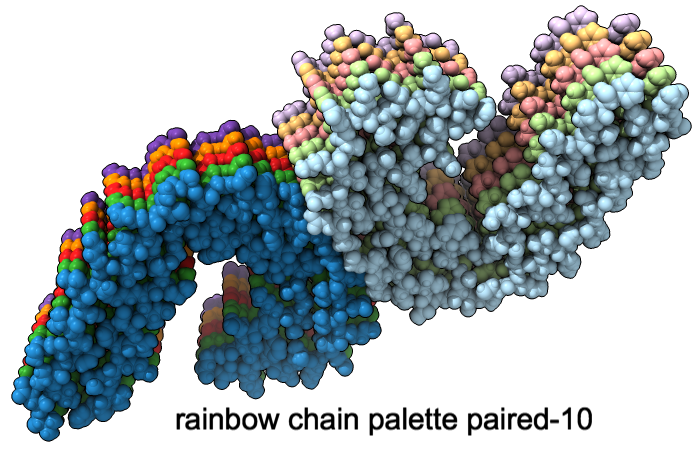
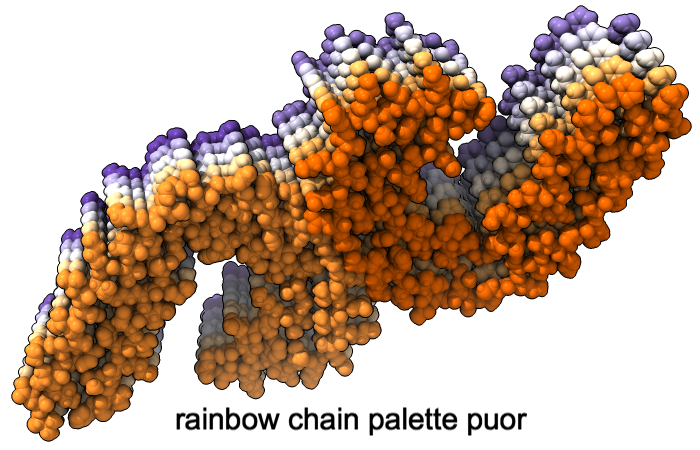
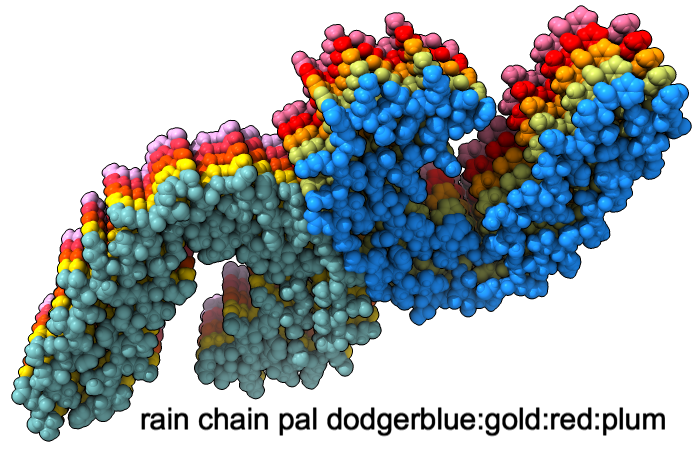
A “palette” or ordered series of colors is used to color items sequentially (rainbow) or by values such as density. The ten chains in PDB 5o3l (paired tau filament) have been colored with the commands shown as 2D labels in the images. The first two examples at left use predefined palettes (credit to www.ColorBrewer.org, color specifications and designs by Cynthia A. Brewer, Pennsylvania State University), whereas the third shows specifying colors individually.
 More features...
More features...
Example Image
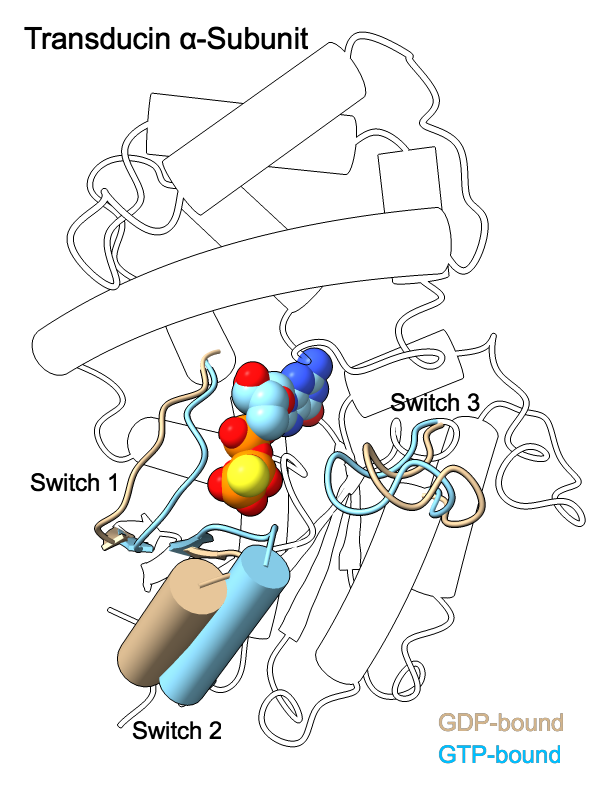
The GDP- and GTP-bound conformations of the transducin α-subunit (1tag and 1tnd, respectively) differ primarily in three regions, termed switch 1, switch 2, and switch 3. The structures have been superimposed with matchmaker and shown as cartoons, with “empty” outlines where the structures are almost the same (for simplicity, only one conformation's outlines are shown). The GTP analog GTPγS is displayed as spheres color-coded by heteroatom. For 2D labels and image setup other than structure orientation, see the command file switch.cxc.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.