

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
Streptomyces umbrella toxin particles block hyphal growth of competing species. Zhao Q, Bertolli S et al. Nature. 2024 May 2;629(8010):165–173.
Structural basis of Integrator-dependent RNA polymerase II termination. Fianu I, Ochmann M et al. Nature. 2024 May 2;629(8010):219–227.
Molecular insights into capsular polysaccharide secretion. Kuklewicz J, Zimmer J. Nature. 2024 Apr 25;628(8009):901–909.
Mechanical activation opens a lipid-lined pore in OSCA ion channels. Han Y, Zhou Z et al. Nature. 2024 Apr 25;628(8009):910–918.
Bond analysis in meta- and para-substituted thiophenols: overlap descriptors, local mode analysis, and QTAIM. Barbosa WG, Santos-Jr CV et al. J Mol Model. 2024 Apr 19;30(5):139.
More citations...News
January 22, 2024
ChimeraX 1.7.1 is available, with fixes for a few miscellaneous bugs that were identified after the 1.7 release.
December 19, 2023
The ChimeraX 1.7 production release is available! See the change log for what's new. Future Mac releases will require macOS 11 or higher.
November 6, 2023
The ChimeraX 1.7 release candidate is available – please try it and report any issues. See the change log for what's new.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325, Chan Zuckerberg Initiative grant EOSS4-0000000439, and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Feature Highlight
Morphing between atomic structures can be calculated wih the morph command and played back in an animation. This movie shows morphing between two conformations of the FGFR1 kinase domain:
Morphing and other setup was done with the command file kmorph-prep.cxc, followed by interactively positioning the structure and saving the view with the command view name p1 (generally a session would also be saved at this point), then running kmorph-play.cxc to add 2D labels and record the movie.
More features...Example Image
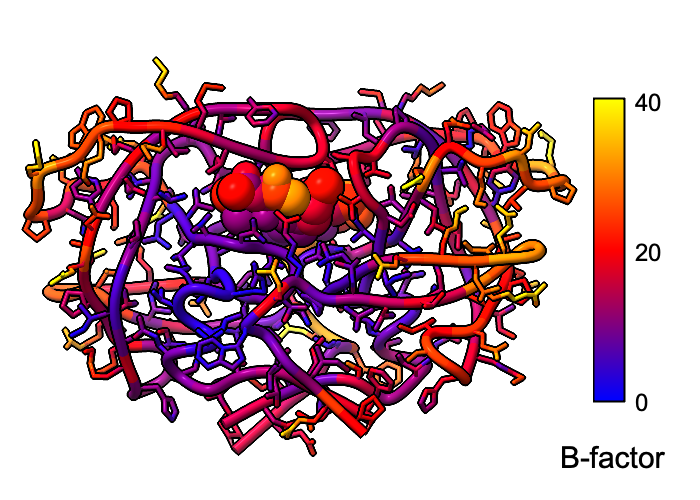
Atomic B-factor values are read from PDB and mmCIF input files and assigned as attributes that can be shown with coloring and used in atom specification. This example shows B-factor variation within a structure of the HIV-1 protease bound to an inhibitor (PDB 4hvp). For complete image setup, including positioning, color key, and label, see the command file bfactor.cxc.
Additional color key examples can be found in tutorials: Coloring by Electrostatic Potential, Coloring by Sequence Conservation
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.