Volume Visualization using Chimera
Tom Goddard
November 8, 2001
NCRR site visit
All images on this page ©2004 The Regents, University of California; all rights reserved.
Introduction
- I've been developing volume display in Chimera to assist several
research projects.
- Volume data means 3D arrays of numbers
- Examples: Electron microscope, light microscope, crystallographic
electron density maps, ...
- Will show 2 Chimera extensions that provide display and path tracing
of volume data.
- Main message of this talk: Chimera is useful for analyzing data over a wide range of resolutions.
Large Range of Data Resolutions
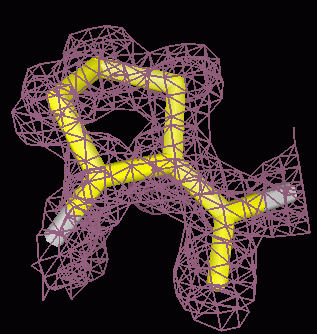
Crystallographic density
|

Electrostatic potential
|

Water occupancy map (1A)
|

Electron cryo-microscopy (7A)
|
Electron microscopy (300A)
|
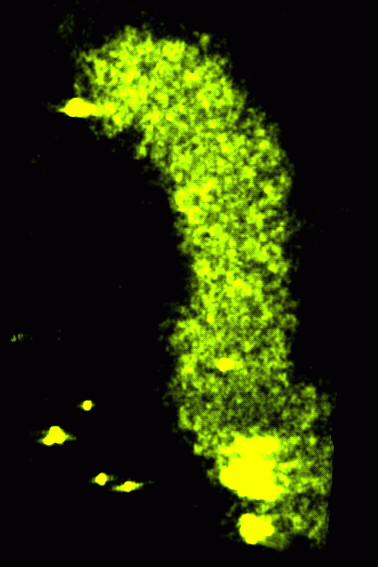
Light microscopy (2000A)
|
Barcoded Chromosomes

|
- Arrangement of chromosomes in the cell nucleus.
- Collaboration with Mike Lowenstein in John Sedat's lab, UCSF.
- 3D light microscope images of chromosome 2L in Drosophila nuclei.
- Can determine whether specific stretches of DNA are attached to the
nuclear envelope.
- Can study spatial arrangements characteristic of differentiated cell types.
- Can study relation between gene transcription and spatial arrangement.
|
Tracing Chromosomes
|

|
- Drosophila chromosome 2L has 13 fluorescently labeled segments using 3 fluorophors.
- Trace path of chromosome by finding expected sequence of color spots
in light microscope data.
- Different colors provide improved resolution
- Colors allow correct interpretation when spots are missing
- Prior to Chimera, tracing was done by flipping through a stack of
2 dimensional images and typing observed sequence of spot id numbers
into a file.
- DAPI stain in light microscope images illuminates all DNA.
- Nuclear envelope can be shown as boundary of DAPI stained region.
|
Low Resolution Protein Structures from
Electron Cryo-Microscopy

|
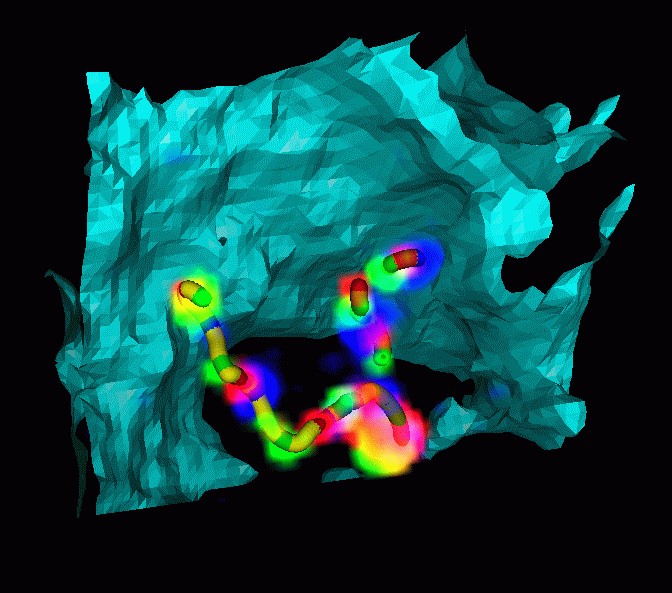
- Rice dwarf virus capsid protein structure determined
- 7 angstrom resolution is sufficient to model alpha helices and beta sheets.
- Single particle cryo-EM can handle large complexes and does not
require crystalization.
- Useful for determining protein folds
- Can determine relative orientations of components of complexes
- Work is done by
Wah Chiu's
group at
Baylor College of Medicine,
National Center for Macromolecular Imaging.
- Alpha helices are found in density map by program
helixhunter
- Helixhunter developed by
Wen Jiang,
Matthew Baker,
in Wah Chiu's lab.
- Display predicted helices superimposed with EM data.
|
Tracing Connections between Helices

|
- Interactively trace turns connecting helices.
- Goal is to find correct helix order, not exact turn structure.
- Helices predicted from amino acid sequence can be matched to those
found in density map.
- Without Chimera, Wah's group did turn tracing on 2 dimensional
images in Photoshop.
- Rice dwarf virus analysis was completed 5 months ago at start of
our collaboration.
- Chimera not used for results published in October issue of
Nature Structural Biology.
- Homology to known crystal structure of Bluetongue virus P7 capsid
protein used in tracing turns.
|
Strengths of Chimera Volume Visualization
- Molecular models, VRML for low resolution models, and volume data
- Show volumes as transparent solids, meshes and surfaces.
- Marker placement and path modeling
- Precomputed subsampling for large data sets
- Subregion selection with mouse, zone selections around markers and atoms
- Python extensions for custom modeling
- 3D image processing, ridge finding, gaussian filtering, ...
Future Development
- Multi-scale models
- Volume movie player with analysis capabilities
- Use Phantom 3D force feedback input device for marker placement and path tracing.
- Flood fill with interactive range control to select or sculpt away
volume regions.
- Non-orthogonal data display for crystallography, accomodate skewing matrix
- Automated marker placement and path tracing.
- Interactive construction of triangulated surfaces using placed markers
How to Depict Macromodels

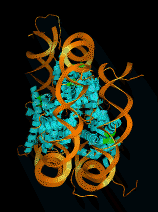
- Ribbon representation of large ribosomal subunit
- Coarser representations are useful for:
- selecting components of interest
- examining component assembly
- moving structure interactively
Analyzing Macromodels: An Example

- How are nucleosomes arranged in the chromatin 30 nm fiber?
- Model of irregular nucleosome placement could come from electron microscopy. (Image shows helical placement with randomized orientations.)
- We have crystal structures of a nucleosome, ~150 bases DNA wrapped around histone core
- Show segment of fiber as aspirin tablets representing nucleosomes
- Limit view to a few nucleosomes and show atomic resolution detail
- Look at how histone tails form inter-nucleosome contacts
Macromodel Development Objectives
- Goal is to provide interactive visualization of models over a wide
range of resolutions.
- Provide low resolution surface representation of macromodel components
- Support data formats for macromodels
- Target systems where we have atomic resolution structures
- Useful for virus capsids, actin / myosin systems, ion channels,
ribosome complexes, collagen bundles, cyto-skeleton, nuclear matrix, ...
Volume Time Series
- Provide volume movie player with analysis capabilities
- Examples from current collaborations:
- Understanding water localization around collagen
- Tracking chromosome transformations during the cell cycle
Water Dynamics around Collagen

- Collagen water localization changes over the course of molecular
dynamics run
- Do preferred water positions vary as collagen bends?
- Are apparent water positions statistical artifacts?
- Look at water occupancy volume movie to guide quantitative analysis.
- Allow comparing 2 or more sections of volume movie simultaneously
with sliders controlling position in movie.
Chromosome Dynamics during the Cell Cycle
- John Sedat's group has made movies of fluorescently labeled chromosomes
in live cells.
- Track motion of labeled segment of chromosome during cell cycle.
- Localization in certain areas may correlate with gene transcription.
- Manual or automatic spot tracing capabilities needed.