Tom Goddard
January 14, 2009
Demonstration for Lincoln High School SMART team which is making a 3d physical model of mevalonate kinase.
PDB models from the Institute for Molecular Manufacturing page of diamondoid molecular machine parts.



This is a nano version of the differential in a car that drives the two rear wheels allowing them to rotate by different amounts when turning the car.
Show diffGear.pdb. Select half and hide atoms. Select individual atoms, use sc shortcut to extend to connected atoms, and da to display. Use move selection mouse mode to move individual parts out for better viewing.

PDB 2r3v. Sphere. Rainbow chain (rc). What are red balls? Water (oxygen = red). Menu Select / Residue / HOH. Menu Actions / Color / Cornflower blue. What species is this protein from? PDB header says Homo Sapiens. Show in text editor.

Why 4 copies of the enzyme? Crystal had about 1000 trillion (~1 mm^3 in size) tiled copies of these 4 shapes. They have slightly different neighbor contacts.
Multiscale 3x3x3 crystal unit cells. Select all (sa). Color to match chain atoms (c=). To see original 4 copies shrink the others. Select one surface, up arrow key, resize mouse mode (sz), drag right mouse button.


Show stack of planes edge on. Drag select plane containing original copies. Hide other chains. Show single crystal plane face on.


PDB header says chains A,B probably form a dimer. "Dimer" means a natural state in cells is 2 copies stuck to each other. How is that determined? Contacting surface area between two copies is large. Experiments are needed to confirm that prediction.


End-to-end pair is dimer. Side-by-side pair is not dimer. Notice contacts are just small patches in side-by-side pair. Like finger tips touching.
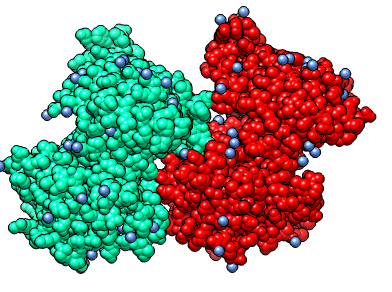
Look at monomer. Select chain A, round ribbon (rr), color by secondary structure (c2), invert selection (is), hide atoms (ha) How many ends of the ribbon? Four. Mouse over shows break 76-79 missing. Why are these missing?

Atom positions were determined by diffracting x-rays through crystal. Example diffraction pattern below is for crystalized rhinovirus.

Density map for PDB 2r3v available from PDB web page. Density map shows all copies of protein. Show density just around chain A. Select chain A. Volume dialog zone, radius 3 Angstroms.
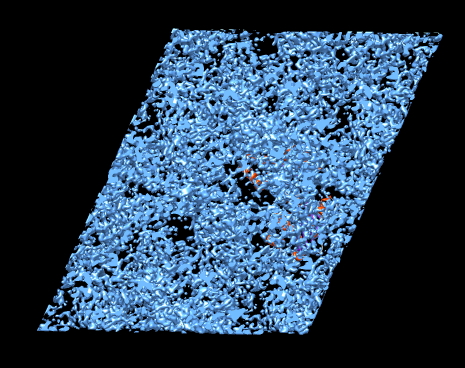

Show map just around break in chain (76-79). Select residues 65-90 (select :65-90.A). Volume dialog zone. Actions / Focus. Radius 5 Angstroms. Mesh. Transparent. Show side chains (da). Loops at surface of protein are sometimes floppy and have different shapes in the many molecules in the crystal smearing out the density.

Structure 2r3v does not have mevalonate bound. Another mevalonate kinase structure 2oi2 has diphosphomevalonate.
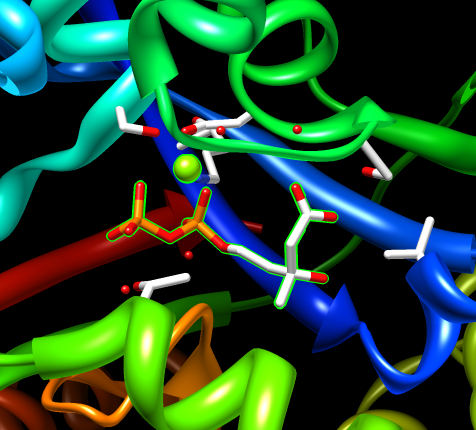
Display clashes. Select chain het. Used Tools / Structure Analysis / Find CLashes, designate selected atoms, vdw overlap 0, continuous update. Use move selection mouse mode to move just mevalonate. Actions / Set Pivot. Use space navigator to move full structure.


If you had a structure without mevalonate you might try to place it by hand.