
| unknown | |
| dipeptide epimerase | |
| chloromuconate cycloisomerase | |
| muconate cycloisomerase (syn) | |
| muconate cycloisomerase (anti) | |
| N-succinylamino acid racemase 2 | |
| o-succinylbenzoate synthase | |
| unclassified |
Demo for NIH site visit November 2011
Updated August 2014 (example results session
mca-5models.py created February 2014)
Elaine Meng, meng [at] cgl.ucsf.edu
This scenario focuses on functional annotation of a protein sequence from the bacterium Methylococcus capsulatus (based on research in the Jacobson, Gerlt, Almo, and Babbitt labs, as described in: Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily, Lukk T et al., Proc Natl Acad Sci USA 109:4122 (2012)).
The sequence is annotated as a chloromuconate cycloisomerase at Genbank (gi 53803900) and a putative chloromuconate cycloisomerase at UniProt (Q607C7). Chloromuconate cycloisomerases are a subset of the enolase superfamily. However, various lines of evidence suggest the unknown is instead a dipeptide epimerase (a different subset of the enolase superfamily) and may have a different substrate specificity from previously well-characterized dipeptide epimerases.
Network, sequence, and structure analysis with RBVI tools can be used to investigate this protein. The sequence, aka MCA 1834:
>tr|Q607C7|Q607C7_METCA Putative chloromuconate cycloisomerase MKIADIQVRTEHFPLTRPYRIAFRSIEEIDNLIVEIRTADGLLGLGAASPERHVTGETLE ACHAALDHDRLGWLMGRDIRTLPRLCRELAERLPAAPAARAALDMALHDLVAQCLGLPLV EILGRAHDSLPTSVTIGIKPVEETLAEAREHLALGFRVLKVKLCGDEEQDFERLRRLHET LAGRAVVRVDPNQSYDRDGLLRLDRLVQELGIEFIEQPFPAGRTDWLRALPKAIRRRIAA DESLLGPADAFALAAPPAACGIFNIKLMKCGGLAPARRIATIAETAGIDLMWGCMDESRI SIAAALHAALACPATRYLDLDGSFDLARDVAEGGFILEDGRLRVTERPGLGLVYPD
Sequence searches with MCA 1834 and incorporation into similarity networks suggest that it belongs to the enolase superfamily like the chloromuconate cycloisomerases, but instead groups much more closely with the dipeptide epimerases. Some of the original network analysis is illustrated in Fig 1 of the paper: a sequence similarity network of known and putative dipeptide epimerases, in which MCA 1834 is one of two magenta squares.
The network image below shows the “unknown” MCA 1834 as a yellow rectangle along with part of the enolase superfamily. You can see that the unknown clusters more with the dipeptide epimerases (light green) than with the chloromuconate cycloisomerases (light red; this was the function suggested by the annotations) or other families in the image.
 |
|
Although the most well-characterized dipeptide epimerases have Ala-Glu specificity (for bacterial cell wall processing), substantial diversity of the family and presence in nonbacterial organisms suggest some members have different specificities.
SFLD protein similarity network XGMML files for analysis in Cytoscape can be downloaded from the SFLD website (more about SFLD networks...).
This scenario shows how the SFLD and Chimera can be used together on a functional annotation problem. Again, we are using these scenarios to give you a small sample of the existing features and how they integrate, not to present new science. The networks mentioned above give a broad perspective on how proteins may relate to one another. To explore sequences and structures in more detail, I'll use the SFLD website (http://sfld.rbvi.ucsf.edu) and Chimera.
| SFLD homepage | SFLD HMM hits |
 |
 |
Show SFLD home page and then search by enzyme. (Chimera started, mca.fasta copied into text buffer.) Paste sequence into browser, search HMMs... best hit is “dipeptide epimerase” family (~ e-80), followed by the subgroup containing that family (muconate cycloisomerase), then the superfamily containing them (enolase), then three other families in the same subgroup including the currently annotated function, chloromuconate cycloisomerase.
Could get alignment with family members from this page, but instead click link to go to the dipeptide epimerase family page (will show alignment later): http://sfld.rbvi.ucsf.edu/django/family/10/
Page contents include links back up the hierarchy, an overall structure image, description of family, enumeration of SFLD contents for that family, an active site image showing family-conserved catalytic residues, and a diagram showing the overall reaction.
The active site image shows the structure of one of the well-characterized dipeptide epimerases in complex with substrate Ala-Glu (PDB 1tkk chain A).
| SFLD family page |
 |
I can click the active site image to open the corresponding session in Chimera (more...). The session was downloaded and opened in Chimera running on this computer. Explain residues: one Lys abstracts a proton from the Glu alpha-carbon (OXT is missing from structure, C-term carboxylate should be shown as interacting with metal), the metal stabilizes the extra negative charge in the intermediate, the other Lys supplies the proton from the other side to invert the carbon center.
In SFLD family page, mention network download, alignment display; choose Align Sequence(s), paste in mca.fasta, choose to view results using Chimera, click Align.
The alignment is shown in the Chimera sequence viewer (Multalign Viewer or “MAV”). Many parts of Chimera open separate dialogs and windows. The HMMer program used for HMM creation and searching puts the query at the bottom. Chimera compares sequences and structures and automatically associates them as appropriate. In this case, the structure associates with gi16078363.
Command: modelcol tan (to make association clearer)These lines above the sequences are called “headers” – I'll hide the ones from HMMer. The Conservation header is calculated in Chimera, and I'll say more about that in a moment.
MAV menu: Edit→Reorder Sequences, move query and struct-assoc seq to top
MAV menu: Preferences→Appearance, change Color scheme to black
MAV menu: Headers, uncheck PP cons, RF (also Consensus if shown)However, these are just the catalytic residues, shared by several other families (i.e. functions) in the enolase superfamily, including the annotated function, chloromuconate cycloisomerase. There is an additional motif that, at least with present knowledge, is diagnostic of dipeptide epimerase activity: a DXD near the the end of the alignment.
Command: sel residues (“residues” is session alias for catalytic residues)
MAV window: selection is green-highlighted; see query has all 5 conserved
MAV window: scroll to locate DXD motif, click-drag to draw box (see figure)
Command: disp sel (then Ctrl-click in empty area of window to clear selection)
Command: ~rlab; focus
| Chimera with Ala-Glu epimerase and family multiple sequence alignment (MSA) |
 |
Besides this motif and the catalytic residues, the dipeptide epimerase HMM is picking up additional signals throughout the alignment. Areas of greater conservation are indicated by higher bars in the Conservation header.
MAV menu: Preferences→Headers, Conservation style AL2CO
(can adjust parameters, see header change)
MAV menu: Structure→Render by Conservation
- Worms: min value radius 0.25, max 1.5, affect no-value true, Apply
- conservation is higher in the active site and core
- to restore ribbon: Worm style non-worm, OK
 |
| Chimera showing conservation with “worms” |
(The following uses pre-defined aliases. To make them available in your own Chimera, save alias.com as plain text and open it in Chimera with menu: File→Open.)
use zone4 or z4 alias (e.g. Command: zone4 or Chimera menu: Aliases→zone4)Alpha-carbons in both the enzyme and substrate are shown as balls. I already mentioned the DXD motif that binds the substrate N-terminus and the interaction of the C-terminal carboxylate with the metal ion. These parts would stay the same; the parts that would be different in different dipeptides would be the sidechains. The substrate Ala sidechain contacts I298 (Ctrl-click any atom in that residue to select), the Glu sidechain forms a salt bridge with R24 (Shift-Ctrl-click any atom in that residue to add it to the selection).
if dim, use white alias (blk alias to reverse)
Command: rlab selThese interactions are described in the paper about this structure (1tkk). I've just been going by eye, but the Chimera tools for identifying H-bonds and other contacts could certainly be applied.
In alignment, scroll to view selected positions: R24 is conserved in the query, but I298 is a negatively charged residue, Asp, in the query. So the first-order guess from sequence is that the sidechain of the substrate N-terminal residue could be polar or even positively charged, while the substrate C-terminal residue could still be glutamate, as in this structure. However, that is a simplistic guess, and it is not obvious from the 2D sequence how the pocket may differ in 3D. A logical next step would be to model the structure of the unknown.
Command: ~sel
Command: ~rlab
Chimera includes an interface to the Modeller program for comparative (homology) modeling and/or refinement, run locally or on a Web service provided by the RBVI. Modeller is developed by the Sali group.
| Chimera-Modeller interface |  |
Comparative modeling can be launched quite easily given the necessary inputs, a target-template sequence alignment and a template structure. In fact, that's what we have now:
MAV menu: Structure→Modeller (homology)(*Academic users can register free of charge to receive a license key. Commercial entities and government research labs, please see Modeller licensing. However, to continue with this demo you could skip to the next paragraph and get the session with example results instead of running Modeller. See also Chimera's ModBase fetch, which does not require a license key.)
- target: Query
- template: the 1tkkA-associated seq (gi|16078363, 30.3% ID)
- enter Modeller license key*
- click OK
This takes 3 or 4 minutes, so I'll step over to the oven and take out the already baked delicious pie... that is, start another Chimera and restore a session saved after the modeling step (mca-5models.py). I'll leave the first one going.
When the models are returned, they are automatically opened in Chimera and superimposed on the template. The models are listed in a dialog along with various quality scores calculated by Modeller, and I can click through to view them individually or together. I won't go into detail about these scores other than to say they are based on statistical potentials. In a real project one would calculate more initial models and carefully select ones to pursue further, possibly performing refinements, but for today's purposes I'll just take the one with the best zDOPE score and close the others. In the mca-5models.py session, the model with the best zDOPE score is #1.5, thus:
Command: close #1.1-4
use zone4 alias
 |
| Chimera with template and 5 models |
Command: sel residuesAs previously noted from the sequence alignment, I298 in the template structure is an aspartic acid in the model (D296). While R24 is conserved, the model contains an additional negatively charged residue in the vicinity (E51). These differences suggest that the preferred substrate may not be a glutamate dipeptide, but possibly something with net positive charge.
MAV menu: Structure→Expand Selection to Columns (one of my favorite features!)
Command: col dark slate gray sel
Command: ~sel
Another thing to look at is the pocket surface, which gives a better sense of its shape and size, and can be colored by various properties.
Command: snocap(again using aliases from alias.com) To view one at a time, show/hide individual surfaces and structures using the S checkboxes in the Model Panel (Chimera menu: Favorites→Model Panel).
Command: surfz 8
Measure and Color Blobs (in Chimera menu under Tools→Surface/Binding Analysis) shows the pocket volume of the model is ~50% greater than that of the template structure. Whereas model #1.5 in the mca-5models.py session has a completely enclosed pocket, this will not necessarily be true of other models.
| 1tkkA (template) | MCA 1834 (model) |
 |
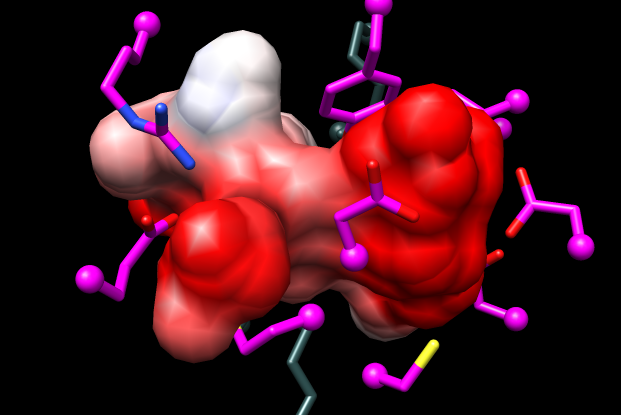 |
Coulombic Surface Coloring (in Chimera menu under Tools→Surface/Binding Analysis) shows the model pocket is more dominantly negative than that of the template (see figure).
The predicted specificity of this protein from the Jacobson group's modeling and docking was for dipeptides with one or both sidechains positively charged, and this was subsequently verified by enzymology and crystallography in the Almo and Gerlt labs. (predicted as Lys-Xxx epimerase, expt/struct gave Pos-Pos specificity; experimental structure 3rit is complex with L-Arg-D-Lys) This project included predicting and experimentally verifying the specificity of not just this protein, but several additional dipeptide epimerases, allowing annotation transfer to >700 sequences.
The newly identified specificities will be added to the SFLD as subfamilies of the dipeptide epimerase family, with associated alignments, HMMs, and network information, similar to what is provided for the higher levels in the hierarchy.
To summarize, I've applied a combination of RBVI tools and resources to this functional annotation problem, including: