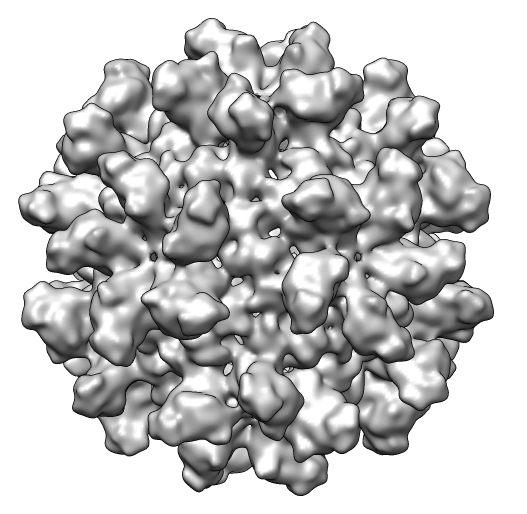
Opened emd_1862.map (#0),
size 2553, grid spacing 1.27 Å
contour level -363, minimum -17270, maximum 14818
mean -6649.5, sd 3176.6, rms 7369.3, 16-bit integer
This is a hand mock-up of proposed Chimera capability to automatically log analysis done in Chimera, including thumbnail images, numeric results and links to sessions, data files, movies, ....
The data file and this web page can be downloaded rsvdata.zip (110 Mbytes). The Chimera session files contain references to electron microscopy map files included in this download. The sessions were made with a Chimera 1.6 pre-release (April 2011).
Thu Apr 7 17:48 PDT 2011
| 1 |
Opened emd_1862.map (#0),
size 2553, grid spacing 1.27 Å | 
|
| 2 |
Opened 1em9.pdb (#1), 2 chains A B, 2257 atoms, x-ray 2.05 Angstroms | 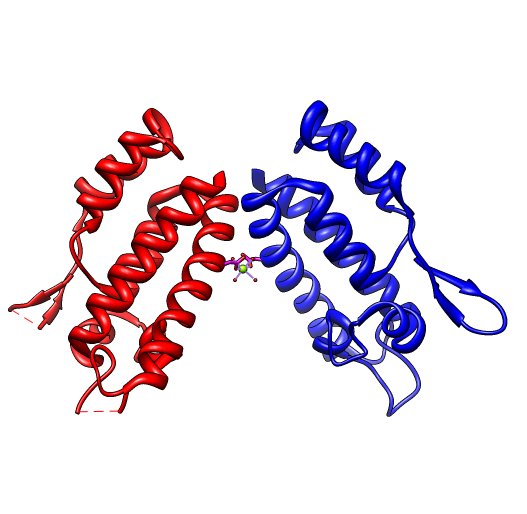
|
| 3 | Deleted 1091 atoms (chain B), 1em9.pdb (#1) | 
|
| 4 |
Opened 1eoq.pdb (#2), 1 chain A, 1209 atoms, NMR | 
|
| 5 | Molecular weight 24.04 KDa, 2375 atoms (1em9.pdb (#1), 1eoq.pdb (#2)). |
| 6 | Contour level 1239, emd_1862.map (#0) |
| 7 | Enclosed volume 1.78e6 Å3, area 454.6e3 Å3, emd_1862.map (#0) |
| 8 | Note: Set contour level to enclose volume 60 * 24 KDa * 1.2 Å3/Da |
| 9 |
Fit 1em9.pdb (#1) in emd_1862.map (#0) | 
| rock movie session9.py |
| 10 |
Segmented emd_1862.map (#0) | 
|
| 11 |
Segmentation grouping emd_1862.map (#0) | 
|
| 12 |
Fit to segment 1eoq.pdb (#2) in region 1966, emd_1862.seg (#3) | 
|
| 13 |
Spin movie 1em9.pdb (#1), 1eoq.pdb (#2), emd_1862.seg (#3) | 
| spin session13.py |
| 14 |
Combined 1em9.pdb (#1), 1eoq.pdb (#2) producing combination (#4) | 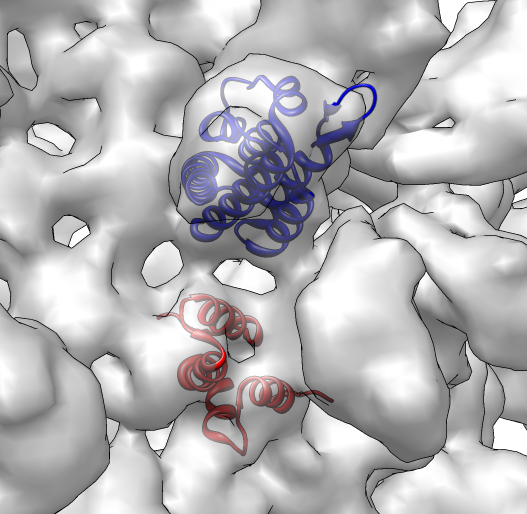
|
| 15 |
Saved ca_fit.pdb (#4) relative to emd_1862.map (#0). |
| 16 |
Symmetry copies ca_fit.pdb (#4) producing (#5.1-59) | 
|
| 17 |
Simulated map ca_fit.pdb (#4), (#5.1-59) producing molmap res 10.4 (#6) | 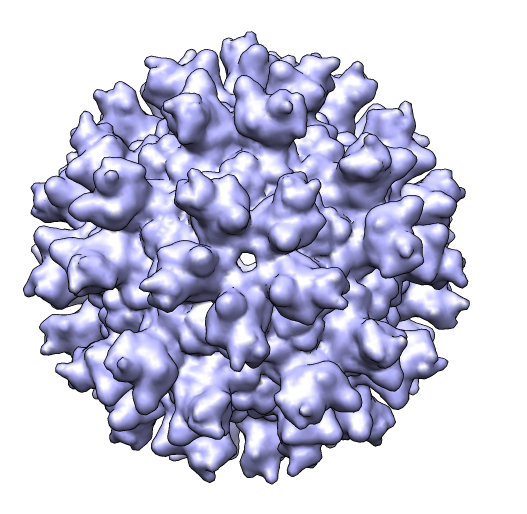
| session17.py |
| 18 |
Saved map ca_fit_60_r10.4.mrc (#6) |
| 19 |
Measure correlation ca_fit_60_r10.4.mrc (#6), emd_1862.map (#0). |
| 20 |
Resampled map ca_fit_60_r10.4.mrc (#6) on grid emd_1862.map (#0) producing (#7). |
| 21 |
Saved map ca_fit.mrc (#7). |
| 22 |
Shifted map emd_1862.map (#0) by 6649 producing (#9). |
| 23 |
Scaled map (#9) by 2.63e-5 producing map (#10). |
| 24 |
Saved map emd_1862_normal.mrc (#10) |
| 25 |
Morph maps emd_1862_normal.mrc (#10) to
ca_fit.mrc (#7) | 
| morph25.mov |
| 26 |
Difference map ca_fit.mrc (#7), emd_1862_normal.mrc (#10). | 
| spin26.mov |
| 27 |
Note: 2 pentamers | 
| session26.py |
| 28 |
Molecular surfaces ca_fit.pdb (#4), (#5.1) | 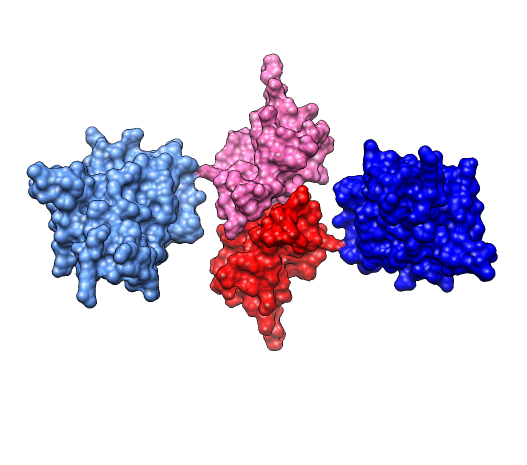
| rock28.mov |
| 29 |
Buried area ca_fit.pdb (#4), (#5.1) |
| 29 |
Buried residues ca_fit.pdb (#4), (#5.1) | 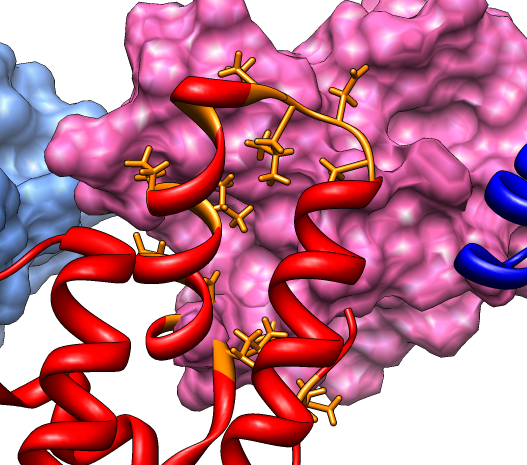
| residue list |
| 30 |
Opened 3p05.pdb (#12), x-ray 2.50 Angstroms | 
|
| 31 |
Fit 3p05.pdb (#12) in emd_1862.map (#0) | 
|
| 32 |
Note: Superposition of HIV and RSV pentamers. | 
| spin32.mov session32.py |
Fri Apr 8 00:15 PDT 2011