Viewing and Analyzing Microscopy Data with Chimera
Tom Goddard
November 15, 2012
Northern California Society for Microscopy
What is UCSF Chimera?
- Molecular visualization and analysis software. Used by a few thousand research labs.
- For analyzing functions of proteins and molecular assemblies.
- For viewing and analyzing 3-dimensional microscopy maps,
mostly electron microscopy.
- Developed at UCSF by the Resource for Biocomputing, Visualization and Informatics, headed by Tom Ferrin.
- Free for academic use. Available on Windows, Mac and Linux.
Examples Imaging Cells and Tissues
Example exotic uses of Chimera molecular visualization software.

| 
|
| Electron tomography of human immune T-cell, 4.6 x 4.6 x 0.17 um.
| Atomic force microscopy of crystallized satellite tobacco mosaic virus,
0.24 x 0.24 um.
|

|  > >
| 
|
| X-ray tomography of Pombe fission yeast cell, 5 x 5 x 12 um.
| Serial block face scanning electron microscopy, rat brain, 50 x 50 x 100 um.
| Focussed ion beam scanning EM, termite gut, 9 x 9.6 x 2.4 um.
|
Example of Molecular Scale Visualization
Here is an example of a common Chimera use: studying molecular structures.
We combine x-ray crystallography protein models with single-particle electron
microscopy of molecular assemblies using map segmentation and fitting and
symmetry. GroEL protein refolding machine, PDB 1grl, EMDB 1080.

| 
| 
| 
| 
|
| GroEL atomic model.
| Hydrophobicity surface.
| EM map of assembly.
| Segment and fit map.
| Full atomic model.
|
Example: Extracting bacteria from termite gut
Bacteria in termite gut convert plant cellolose to useful fuel.
Over 200 species of bacteria in gut. Could some be used for biofuel
production?
|
|
| 
|
Display styles: plane, transparent,
orthogonal planes, box.
| Extract one bacterium.
| 4.4 um long.
Measure length by placing markers on smoothed map.
|

| 
| 
|
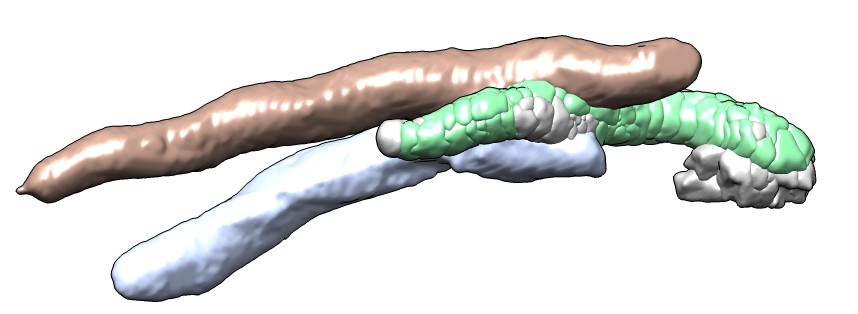
| Another termite gut map.
| Segmented bacteria.
| Autotraced spines.
|

| 
| 
|
| Colored by length.
| Over 6A long.
| Contacts with one bacterium.
|
|
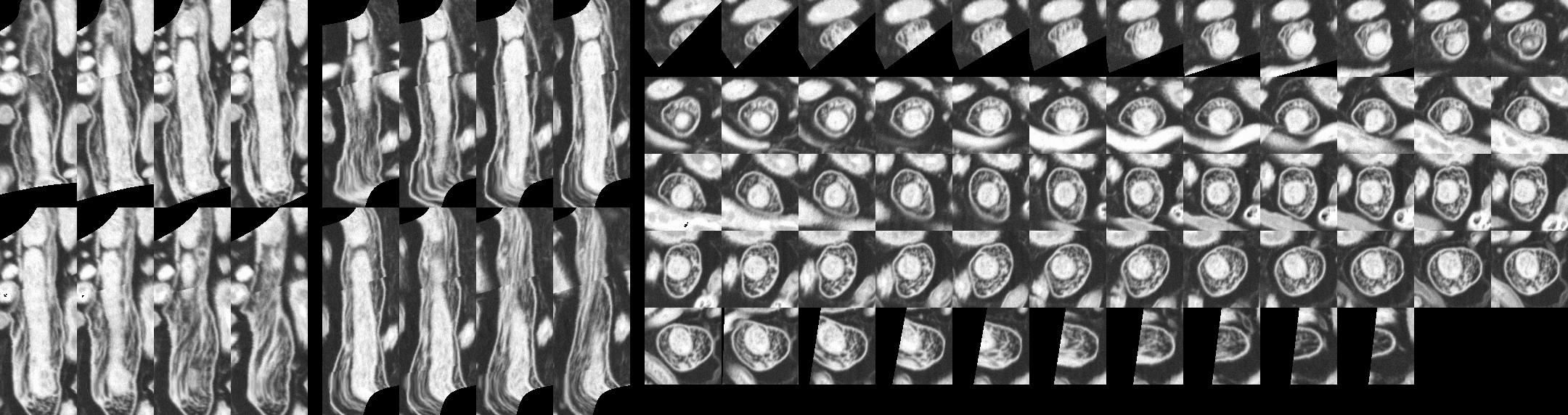
| Cross-sections of one selected bacterium computed from spine path.
|
| 
| 
|
| Segment bacteria by clicking and dragging mouse.
Color spreads to neighboring region but not into already
colored bacteria.
|

Chimera Web Site
Chimera Development Team
Chimera is developed by 5 people
at the University of California, San Francisco.
- Greg Couch - 3d graphics, OpenGL
- Conrad Huang - project leader, web services.
- Elaine Meng - documentation.
- Tom Ferrin - principal investigator.
- Eric Pettersen - structure and sequence analysis.
- Tom Goddard - EM maps and molecular assemblies.






 >
>


















