Chimera Multi-Scale Modeling Extension
Tom Goddard
October 18, 2002
Advisory Committee Talk
All images on this page ©2004 The Regents, University of California; all rights reserved.
Introduction
- For exploring models of large molecular complexes.
- Example systems are viruses and chromosomes.
- Goal is to provide capabilities in Chimera to explore systems where
both large scale and atomic scale structure is important.
- NCRR research project - "Analyzing molecular complexes at multiple resolutions".
- Collaborating with Wah Chiu at Baylor College of Medicine on electron
microscope studies of viruses.
- Collaborating with John Sedat at UCSF on electron and light microscope
studies of chromosomes.
- Will show 4 example systems and demonstrate current Chimera capabilities.

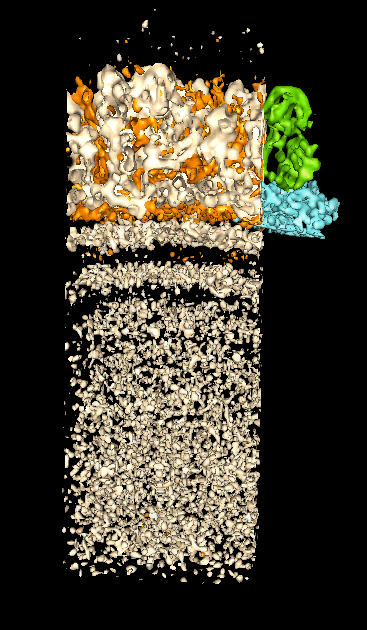
Bluetongue Virus Core Particle
- PDB structure 2btv, 1998
- Picture shows ribbon diagram of one unit of icosahedral particle and
surfaced representation of another unit.
- Making low resolution surfaces is done by multi-scale extension.
- Full particle 700 A diameter, 3.5 A resolution, 1000 crystals,
3 million atoms (no hydrogens)
Bluetongue Virus Architecture
- Multi-scale extension makes 60 copies of unit cell to show virus particle.
- Outer layer has 260 trimers of VP7 protein in 5 symmetry classes.
- Inner layer has 60 dimers of VP3.
- Computer view of virus architecture shown in diagram.
- Not all of this architecture is in computer readable form from PDB entry.
- To hide outer layer computer must know what "outer layer" is.
- Multi-scale extension focuses on encoding and using this hierarchical
structure information.
Why Look at Full Virus?
- Image shows viral dsRNA attached to outside of virus particle.
- Thought to be a means to sequester excess viral RNA to avoid cell shutdown.
- Atomic scale and large scale useful for studying stickiness of shell and RNA.
- 60% of RNA / trimer contacts are with 5 polar residues.
- Work published by David Stuart's group at Oxford, September 2002.

Demonstration Showing Bluetongue Virus in Chimera
- Show selection of outer monomer with mouse.
- Show selection promotion to trimer and trimer class.
- Show hiding trimers around 5 fold axis.
- Hide outer layer.
- Show all except half of outer layer.
- Show inner layer monomer as ribbon.
- Show other inner layer monomer as ball and stick colored by element.
- Use near and far slice planes to show thinness of inner layer.
Current Multi-Scale Capabilities
- Creates low resolution surface depictions.
- Creates multimeric complexes from monomers.
- Understands higher order structure.
- Allows both atomic and surface display styles.
- Details:
- Models made automatically from PDB chains and matrix transformations.
- More control specifying higher levels of structure using Python script.
- Select chains with mouse and promote selections.
- Selections control what is displayed,
display style (surface, ribbon, ...), and colors.
- Adjustable level of detail effects all surfaces.
Rice Dwarf Virus Electron Microscope Density Map
- Rice dwarf virus is in the same family as Bluetongue virus (reoviridae).
- Core particle has 2 layer shell and dsRNA genome.
- Crystal structures of shell proteins have not been determined.
- Density map from electron cryo-microscopy of 3261 full virus particles
from Wah Chiu's lab.
- Density map is hand segmented into layers, trimers, dimers and monomers.
- Averaging outer layer P8 monomer EM density gives 7 A resolution.
- Secondary structure (alpha helices and beta sheets) visible.
- Pictures: full particle, inner layer dimer, shell and dsRNA strata,
outer layer trimer, outer layer monomer.
Heterogeneous Data in Multi-Scale Models
- Multi-scale model can consist of components that are segmented regions
of EM density map.
- Modeled backbone trace can also be a component.
- Structure components can contain atomic models, X-ray density maps,
EM density maps, secondary structure tracings, theoretical models
(eg. dsRNA genome packing).
- A multi-scale model component can contain more than one type of data.
Virus Images from Other Software
- Journal images of viruses commonly done in GRASP, MolScript, IRIS Explorer, Raster3D, ....
- These systems provide beautiful high resolution publication images.
- Radially colored density maps of viruses are common.
- The graphics are typically molecular surfaces that render too slowly for
use in interactive analysis.
- These programs do not understand the different structural levels of
large complexes which limits their usefulness for interactive analysis.
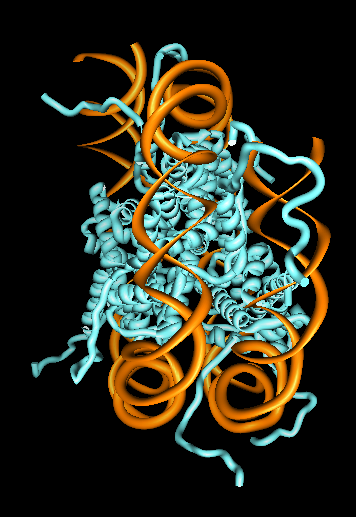
Chromatin 30 nm Fiber
- DNA in eukaryotes is usually packaged as a 30 nm fiber
called chromatin.
- Fiber is composed of nucleosomes and accessory proteins.
- Nucleosomes consist of a histone protein octamer wrapped with
~2 turns of DNA, about 200 base pairs.
- Nucleosome crystal structure is shown.
- Conceptual model of fiber with 100 nucleosomes is shown.
Detailed structure is unknown.
- Histone tails thought to hold chromatin together.
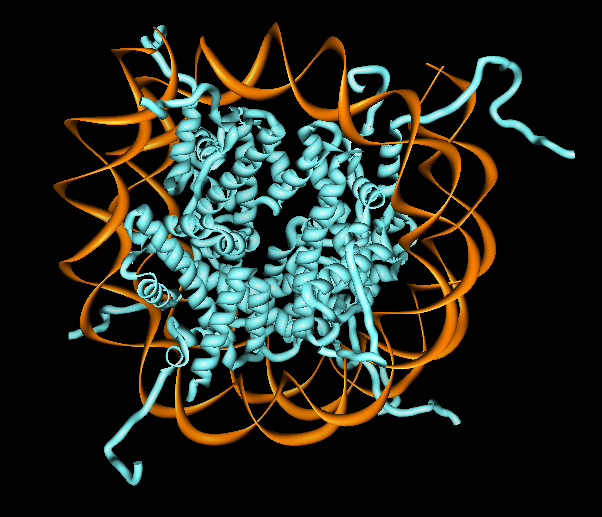
Displaying Less Detail
- Showing nucleosomes as pellets provides
clearer view of chromatin model than more detailed rendering.
- User specified geometric representations could be supported.
- A single user specified triangle could represent a virus shell trimer.
- Need control over which level of structure hierarchy gets surfaced.
- Surface resolution needs to be adjustable to not overwhelm graphics
hardware.
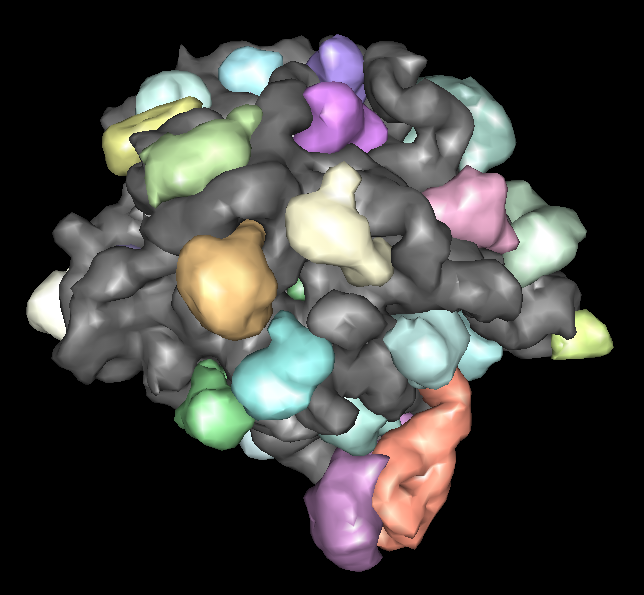
Large Ribosomal Subunit
- PDB model 1jj2, 30 different chains, 90000 atoms (no hydrogens).
- Non-multimeric complex.
- There are 8 large ribosomal subunit structures and several other
partial structures in PDB.
- Different structures have bound anti-biotics, or protein synthesis intermediates, or are from different species.
- Low-resolution display capability can be useful to compare these
large complexes.
Future Multi-Scale Improvements
- Optimization. Currently 10 times slower surfacing than necessary.
- User interface to specify model hierarchy.
- File format to save model hierarchy.
- Handle EM or X-ray density maps or secondary structure models
(currently only molecular chains).
- Surface representation for model components above chain level.
- Surface level of detail for individual components.
- Naming of different levels of model ("outer layer", "VP7 trimer", ...).
- User specified representations (cylinders, individual triangles).
- Write documentation.
- Interact with leading edge users to guide development.
- Distribute with Chimera.