Tom Goddard
November 17, 2008
Representation of viruses in the remediated PDB archive.
Lawson CL, Dutta S, Westbrook JD, Henrick K, Berman HM.
Acta Crystallogr D Biol Crystallogr. 2008 Aug;D64(Pt 8):874-82. Epub 2008 Jul 17.

Paramecium bursaria chorella virus type 1 (PBCV-1) is PDB entry 1m4x.
Has 3 chains A,B,C forming a trimer.
1680 trimers for the capsid.
# loop_ _pdbx_struct_assembly.id _pdbx_struct_assembly.details 1 'complete icosahedral assembly' 2 'icosahedral asymmetric unit' 3 'icosahedral pentamer' 4 'icosahedral 23 hexamer' 5 'pentasymmetron capsid unit' 6 'trisymmetron capsid unit' PAU 'icosahedral asymmetric unit, std point frame' # loop_ _pdbx_struct_assembly_gen.assembly_id _pdbx_struct_assembly_gen.oper_expression _pdbx_struct_assembly_gen.asym_id_list _pdbx_struct_assembly_gen.entity_inst_id 1 (1-60)(61-88) A,B,C . 2 (61-88) A,B,C . 3 (1-5)(61-88) A,B,C . 4 (1,2,6,10,23,24)(61-88) A,B,C . 5 (1-5)(63-68) A,B,C . 6 (1,10,23)(61,62,69-88) A,B,C . PAU (P)(61-88) A,B,C . # loop_ _pdbx_struct_oper_list.id _pdbx_struct_oper_list.type _pdbx_struct_oper_list.matrix[1][1] _pdbx_struct_oper_list.matrix[1][2] _pdbx_struct_oper_list.matrix[1][3] _pdbx_struct_oper_list.vector[1] _pdbx_struct_oper_list.matrix[2][1] _pdbx_struct_oper_list.matrix[2][2] _pdbx_struct_oper_list.matrix[2][3] _pdbx_struct_oper_list.vector[2] _pdbx_struct_oper_list.matrix[3][1] _pdbx_struct_oper_list.matrix[3][2] _pdbx_struct_oper_list.matrix[3][3] _pdbx_struct_oper_list.vector[3] P 'transform to point frame' 0.30901699 -0.50000000 0.80901699 0.00000 0.80901699 -0.30901699 -0.50000000 0.00000 0.50000000 0.80901699 0.30901699 -0.00000 1 'point symmetry operation' 1.00000000 0.00000000 0.00000000 0.00000 0.00000000 1.00000000 0.00000000 0.00000 0.00000000 0.00000000 1.00000000 0.00000 2 'point symmetry operation' 0.80901699 0.30901699 0.50000000 0.00000 0.30901699 0.50000000 -0.80901699 0.00000 -0.50000000 0.80901699 0.30901699 0.00000 ... 60 'point symmetry operation' -0.50000000 -0.80901699 -0.30901699 0.00000 0.80901699 -0.30901699 -0.50000000 0.00000 0.30901699 -0.50000000 0.80901699 0.00000 61 'build point asymmetric unit' 1.00000 0.00000 0.00000 0.0 0.00000 1.00000 0.00000 0.0 0.00000 0.00000 1.00000 0.0 62 'build point asymmetric unit' 0.999773 0.014247 -0.015805 -49.486 -0.014842 0.999161 -0.038166 14.993 0.015248 0.038393 0.999147 32.902 ... 88 'build point asymmetric unit' 0.967112 0.161474 -0.196522 -130.071 -0.188316 0.973929 -0.126494 95.792 0.170973 0.159342 0.972305 -1.525 # loop_ _pdbx_struct_legacy_oper_list.id _pdbx_struct_legacy_oper_list.name _pdbx_struct_legacy_oper_list.matrix[1][1] _pdbx_struct_legacy_oper_list.matrix[1][2] _pdbx_struct_legacy_oper_list.matrix[1][3] _pdbx_struct_legacy_oper_list.vector[1] _pdbx_struct_legacy_oper_list.matrix[2][1] _pdbx_struct_legacy_oper_list.matrix[2][2] _pdbx_struct_legacy_oper_list.matrix[2][3] _pdbx_struct_legacy_oper_list.vector[2] _pdbx_struct_legacy_oper_list.matrix[3][1] _pdbx_struct_legacy_oper_list.matrix[3][2] _pdbx_struct_legacy_oper_list.matrix[3][3] _pdbx_struct_legacy_oper_list.vector[3] 1 1x61 1.00000000 0.00000000 0.00000000 0.00000 0.00000000 1.00000000 0.00000000 0.00000 0.00000000 0.00000000 1.00000000 0.00000 2 1x62 0.99977300 0.01424700 -0.01580500 -49.48600 -0.01484200 0.99916100 -0.03816600 14.99300 0.01524800 0.03839300 0.99914700 32.90200 ... 1679 60x87 -0.28149639 -0.95353767 0.10734706 -20.06292 0.76749273 -0.29088592 -0.57126126 -188.26516 0.57594612 -0.07842024 0.81371768 -71.92725 1680 60x88 -0.38403872 -0.91790149 -0.09986197 -11.99060 0.75511638 -0.24999640 -0.60605334 -134.06850 0.53133210 -0.30815591 0.78912963 -89.32390
REMARK 300 REMARK 300 BIOMOLECULE: 1 REMARK 300 THIS ENTRY CONTAINS THE UNIQUE NON-CRYSTALLOGRAPHIC REPEAT REMARK 300 UNIT, WHICH CONSISTS OF 3 CHAIN(S). SEE REMARK 350 FOR REMARK 300 INFORMATION ON GENERATING THE BIOLOGICAL MOLECULE(S). REMARK 300 THE ASSEMBLY REPRESENTED IN THIS ENTRY HAS REGULAR REMARK 300 ICOSAHEDRAL POINT SYMMETRY (SCHOENFLIES SYMBOL = I). REMARK 350 REMARK 350 GENERATING THE BIOMOLECULE REMARK 350 COORDINATES FOR A COMPLETE MULTIMER REPRESENTING THE KNOWN REMARK 350 BIOLOGICALLY SIGNIFICANT OLIGOMERIZATION STATE OF THE REMARK 350 MOLECULE CAN BE GENERATED BY APPLYING BIOMT TRANSFORMATIONS REMARK 350 GIVEN BELOW. BOTH NON-CRYSTALLOGRAPHIC AND REMARK 350 CRYSTALLOGRAPHIC OPERATIONS ARE GIVEN. REMARK 350 REMARK 350 BIOMOLECULE: 1 REMARK 350 APPLY THE FOLLOWING TO CHAINS: A, B, C REMARK 350 BIOMT1 1 1.000000 0.000000 0.000000 0.00000 REMARK 350 BIOMT2 1 0.000000 1.000000 0.000000 0.00000 REMARK 350 BIOMT3 1 0.000000 0.000000 1.000000 0.00000 REMARK 350 BIOMT1 2 0.999773 0.014247 -0.015805 -49.48600 REMARK 350 BIOMT2 2 -0.014842 0.999161 -0.038166 14.99300 REMARK 350 BIOMT3 2 0.015248 0.038393 0.999147 32.90200 ... REMARK 350 BIOMT1 1680 -0.384039 -0.917901 -0.099862 -11.99060 REMARK 350 BIOMT2 1680 0.755116 -0.249996 -0.606053 -134.06850 REMARK 350 BIOMT3 1680 0.531332 -0.308156 0.789130 -89.32390
3D complex: a structural classification of protein complexes.
Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA.
PLoS Comput Biol. 2006 Nov 17;2(11):e155. Epub 2006 Oct 5.
More than half of RCSB structures are homo- or heteromeric protein complexes.
Chimera does not recognize and identify identical or similar chains in multi-chain complexes.
Listing all unique contact interfaces between chains would be useful.


As of Nov 12 2008 there are 52444 entries loaded with the following exceptions:
nucleic acid and nuclei acid/protein complexes excluded all virus coat assemblies related entries excluded.
1bbs 1cde 1cyc 1hrb 1lbt 1xys 2cha 2lgs 1bcf
1gto 1xim 2xim 3pcn 3pca 3pcf 3pch 3pci 3pcj
3pck 3pcl 3pcm 3xim 1bcf 2lgs 123d 1ruo 1wio
1wip 3tra
The above entries are available via the PDBidcode search mechanism.
---------------------------------------------------------
Oligomer Number Number Number Examples
size generated homo- hetero-
(accepted) (accepted) (accepted)
---------------------------------------------------------
monomer/complex 19491
dimer 17903 (16472) 13705 (12363) 4194 (4109)
trimer 3554 (3496) 1664 (1611) 1890 (1885)
tetramer 6013 (5962) 3823 (3785) 2190 (2177)
pentamer 398 (394) 195 (195) 203 (199) 1BOV (homo) 1A2K (het)
hexamer 1862 (1845) 1158 (1143) 704 (702) 1AEI (homo) 1AFV (het)
heptamer 100 (99) 48 (48) 52 (51) 7AHL (homo) 1LTI (het)
octamer 786 (779) 453 (450) 333 (329) 1AHU (homo) 1BFV (het)
nonamer 88 (88) 11 (11) 77 (77) 1FWA
decamer 157 (157) 91 (91) 66 (66) 1GTP
undecamer 17 (17) 14 (14) 3 (3) 1WAP
dodecamer 472 (472) 215 (215) 257 (257) 1COA (homo) 1IZB (het)
tetradecamer 46 (46) 34 (34) 12 (12) 1GRL (homo) 1AVO (het)
hexadecamer 98 (98) 16 (16) 82 (82) 2HLC (homo) 1AA1 (het)
octadecamer 27 (26) 6 (6) 21 (20) 1KZU
21meric 41 (41) 0 (0) 41 (41) 1AON (homo)
24meric 144 (140) 85 (85) 59 (55) 1AEW (homo) 3PCA (het)
26meric 14 (14) 0 (0) 14 (14) 1OCC
28meric 21 (21) 1 (1) 20 (20) 1PMA
---------------------------------------------------------
Desmosomes from a structural perspective.
Stokes DL.
Curr Opin Cell Biol. 2007 Oct;19(5):565-71. Epub 2007 Oct 22. Review.

June 2006 EM tomography example fitting HIV gp120/gp41 spikes using Chimera. Spikes recognize target cells and initiate viral fusion. Illustrates combined use of tomography and structure averaging approach.
Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH.
Distribution and three-dimensional structure of AIDS virus envelope spikes.
Nature. 2006 Jun 15;441(7095):847-52. Epub 2006 May 24.

| 
|

| 
| 
|
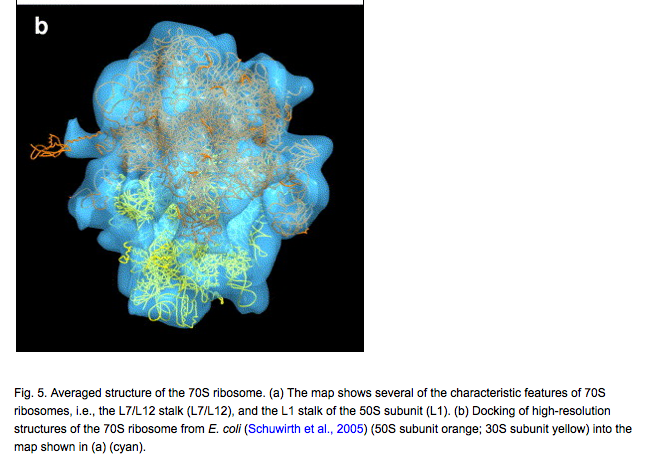
Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition.
Ortiz JO, F�rster F, K�rner J, Linaroudis AA, Baumeister W.
J Struct Biol. 2006 Nov;156(2):334-41. Epub 2006 Jun 3