Human T-cell Chimera Demonstration Notes
Tom Goddard
December 13, 2006.
Presented at Workshop for Visualization of Cellular Complexity
Max Planck Institute, Martinsried, Germany.
This demonstration shows electron microscope tomography of a human immune
system cell in contact with target cell. Vesicles containing serine proteases
are transported to the cell membrane along microtubles to kill the target cell.
The demonstration shows the tomography map, a segmentation of the tomography,
a high resolution microtubule map, and a microtubule atomic model. The
demonstration illustrates use of several Chimera tools: map display,
single plane display, placing markers on maps, segmentation display,
tracing paths, fitting maps in other maps, fitting models in maps,
multimeric atomic model display, display of inter-molecular atomic contacts.
Demonstration
Step by step notes for demonstrating visualization form cellular scale
to atomic scale.
Display cellular tomography and segmentation

- Resize Chimera window to use 2/3 of screen.
- Open map ctl.nc (ctl = cytotoxic T lymphocyte).
- Press Show button on volume viewer dialog.
- Switch surface display to solid. Tomography typically viewed with
volumetric (solid) rendering.
- Adjust transfer function. Lower threshold to 110.
- Note map size (2048, 2048, 76) and subsampling every 8th voxel along
each axis.
- Tilt map to show thickness.
- Start volume planes tool (Tools / Volume Data menu). Obtained from
experimental features web page. Place slider under main window, resize
to equal width.
- Adjust transfer function. Middle node to 150, full brightness.
Adjustment needed since only one plane being displayed.
- Press Play button on volume planes dialog. Adjust step size to 4 to
flip through planes quickly.
- Point out two cells, boundary between them, lytic granules (bright white
blobs). Explain granules are delivered to target cell to initiate apoptosis,
transported along microtubules.
- Open segmentation made in IMOD, ctl.imod.
- Reorient view from t-cell side, looking down 45 degrees, zoom in.
- Blue lytic granules, red microtubules, yellow centriole organized
microtubules, green golgi, white cell boundary.
- Change color of cell boundary so it is different from map. Select it
with mouse. Shortcut co to color it orange.
- Note Chimera cannot create these segmentations and has few capabilities
to use them, other than looking at them.
- Components can be hidden. Select golgi and hide with hs, reshow with sf.
- Microtubules are all one component in IMOD segmentation so they cannot
be hidden or recolored separately. Splitting by connectedness could be added
in the future.
- Chimera has tool to measure volume within surfaces. Will not work on lytic
granules because of open ends at top and bottom. Again could be made to compute
volume even with open ends.
- Just starting to develop Chimera tomography tools.

Trace microtubule
- Trace individual microtubule.
- Hide segmentation using Model Panel.
- Reorient to standard orientation.
- Crop using volume subregion selection central box containing centriole
and golgi in order to flip through planes quickly at full resolution.
- Find favorite microtubule, close to perpendicular to z planes. In plane
20 it is above brightest spot near golgi.
- Zoom in close and center in window.
- Flip through planes 20 - 62 showing its path.
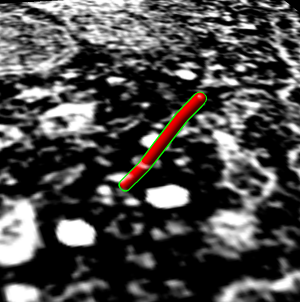
- Start at plane 20 and place marker using volume path tracer. Turn on
drop markers on empty space.
- Resize marker to 50A radius (10nm diameter versus tubule 25nm diameter).
- Color marker red.
- Set center of rotation with cr or Actions / Set Pivot.
- Tilt plane to show that marker is on the plane, then tilt back to face on.
- Place 3 more markers at on higher planes up to about plane 62.
- Drag select all and change link color to red, radius to 50.
- Tilt to about 45 degrees and flip through planes.
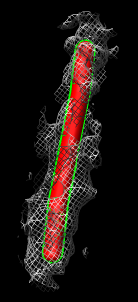
- Show zone around trace, 200A (20nm) radius. Mesh.
- Turn on square mesh or have it saved as default volume dialog setting.
Compare single particle microtubule map to tomogram microtubule
- Compare to single particle microtubule map.
- Open microtubule.mrc. 8A map blurred with 10A Gaussian convolution
because we just want to see tubulin organization.
- Map opens far out of view. Center in view with Cm.
- Color cyan, lower contour level to about 120, surface style.
- Close marker set in path tracer dialog (File / Close marker set),
turn off marker mouse mode and close dialog.
- Hide ctl.nc with model panel.
- Zoom in and orient vertically microtubule.mrc.
- Explain architecture of 13 lengthwise protofilaments, each made of
alpha and beta tubulin monomers (one per bulge) alternating.
- Fit microtubule map in tomography map. Explain that this is just to
illustrate fitting tool. Since protofilaments and monomers not visible in
tomography we won't get any filament orientation info.
- Show ctl.nc, orient vertically, freeze it, and hand
align microtubule map in middle of ctl microtubule, unfreeze,
and show from different angles.
- Show fit map in map tool. Menu Tools / Volume Data. Choose microtubule
as first map. Set microtubule step to 2 to make fit very fast -- does not
effect observed map features. Press Fit.
- Sometimes map is thrown far away (is using data outside zone). If so,
try again near center of ctl microtubule.
- Note correlation of 0.02, essentially 0 because only data within microtubule
contour level used. So correlation measures protofilament and monomer
structure.
- Turn off "use only data above contour level" and press Correlation button
to show slightly higher correlation 0.13 due to tomography having hole in
middle.
Fit tubulin atomic into microtubule map

- Now look at higher resolution. Fit tubulin atomic model in map.
This will show some features for working with single particle maps.
- Hide ctl.nc
- Open tubulin dimer 1jff.
- Opens far out of view, center with Cm.
- Hide microtubule.mrc if it block view of 1jff.
- Color alpha and beta tubulin differently, rc (rainbow chain).
- Switch to ribbon display rr, hide atoms ha.
- Show microtubule.mrc as mesh, step size 1.
- Hand position dimer in map, freezing other models with model panel.
- Select all atoms sa, and focus fo, rotate to show front and back
of hand fit.
- Use fit model in map, menu Tools / Volume Data, press Fit.
- Note that model moved to local maximum average density at atom positions.
- Have undo/redo move on toolbar and show original and final position.
- Note average map value. Should be 145.5 for correct orientation with
longest diagonal helix on outer side of tubule. That agrees with published
fit. Get 144.3 for upside down
orientation. Reorient and try again to get 145.5 -- look for long helix on
outside.
- Note that average map value has no absolute meaning since map is not
normalized, but can be used compare alternate orientations.
- Note number of atoms outside displayed contour is another measure of
quality of fit.
- Clear selection for simpler view and rotate to show inside and outside
of tubule.
Microtubule atomic model

- Now we'll look at atomic contacts between protofilaments.
- Show multimeric form "bu".
- Color cyan microtubule mesh transparent gray roughly (.7,.7.,.7,.7) to
contrast with cyan multiscale surface.
- Reset clip planes and zoom "va" and center of rotation "cr".
- Show original dimer as ribbon. Press multiscale select chains with
loaded atoms, then press style Show... / Ribbon.
- Explain that blue and yellow surfaces depict other alpha and beta
tubulin monomers.
They are low resolution surface calculated from 1jff PDB model.
- Select yellow monomer, then all copies, then change resolution to 3A.
- Explain that Chimera placed them using symmetry matrices to put into 1jff
PDB file (with text editor). Symmetry matrices were from known symmetry of
map (40.6A tubulin monomer spacing, one helical turn of tubulin monomers
rises 3 monomers).
- Note that microtubule has a seam between two protofilaments.
Replace ribbons with surface.
Spin models about vertical axis to follow one yellow helical turn to see
that it hits blue monomer. Contacts between adjacent protofilaments are
usually alpha tubulin with alpha-tubulin and beta with beta. But at seam
it is alpha with beta. (Alpha and beta have 40% sequence identity.)
Atomic contacts between protofilaments
- Now look at contacts between adjacent protofilaments. Look at alpha-alpha
and beta-beta contacts.
- Hide microtubule mesh.
- Zoom in on ribbon and non-seam protofilament neighbor.
- Select one residue of ribbon and up-arrow twice to select full dimer.
Shift ctrl click to add adjacent alpha+beta monomer surfaces to selection.
- Hide other chains using multiscale Hide button, show selected as ribbon.
- Make ribbons match surface colors with multiscale color Ribbons button.
- Reselect full ribbon dimer, switch on "load atoms" in multiscale and
press Contacts button.
- Press up-arrow to select whole residues, and display atoms "da",
show as sticks "st".
- Zoom in on one set of contacts and place mouse over a few residues to
get pop-up balloons identifying residue numbers.


More Chimera capabilities
- Chimera has many more capabilities for studying atomic models. For
example it could compute a sequence alignment of alpha and beta tubulin
and a corresponding structure alignment.
- We are developing Chimera to provide visualization and basic interactive
analysis for length scales from molecules to cells.
- Tomography capabilities are currently much more limited than atomic
model capabilities.
- Volume plane display and IMOD segmentation display are new and not
yet part of the standard Chimera distribution. Available as plugins that
can be downloaded from the Chimera
Experimental Features web page. Show this web page.
Notes
Tomography NetCDF file.
The original tomography MRC map header claims it is signed 8-bit data.
It is in fact 8-bit unsigned. Map reinterpretted as unsigned and inverted
(0 becomes 255, 255 becomes 0) was saved in a NetCDF file ctl.nc. That
file also contains subsampled copies of the map with step size 2 and 4.
Those were created with the Chimera VolumeProcessing/download.py script,
opened as precomputed subsamples with volume viewer and then saved in the
netcdf format. The subsamples allow quick display of the full map without
having to read full map (~300 Mbytes) from disk.
Fitting with symmetry.
Plugin allows fitting one copy of a molecule (e.g. tubulin dimer) by hand
or with local optimization while other symmetric copies automatically
update their positions. Good for seeing clashes.
Clash detection.
Have atomic clash detection that can be used during fitting. Can show
lines between molecule being fit and neighbors when atoms are close.
Real-time update is currently (1.2382) slow even for two tubulin dimers
(~6000 atoms x 2) because calculation is in Python.
Data sets
Cellular tomography.
EM tomography
(EMDB 1273)
of stained plastic fixed human cytotoxic T lymphocyte in
contact with a target cell. Examined transport of lytic granules to
synapse along microtubules.
Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM.
Centrosome polarization delivers secretory granules to the immunological synapse.
Nature. 2006 Sep 28;443(7110):462-5.
Microtuble density map.
Microtubule single particle cryoEM construction to 8A resolution.
(Obtained from KH Downing, not in EMDB as of May 2007).
Publication proposes that single particle reconstruction is better than helical
reconstruction for this data. Rises 3 monomer units in one turn in
left-handed helix. This is 1.5 dimers and implies that the filament
has a seam -- ie is not strictly helically symmetric unless alpha and
beta monomers are considered identical. 13 protofilaments parallel
the microtubule axis. At alpha and beta tubulin have 41% sequence
identity and are essentially indistinguishable at 8A resolution.
Microtububule is about 25 nm in diameter. Symmetry: z-translation
40.6 A monomer to monomer, rotation by angle 360/13 (~= 27.7 degrees)
and translation by 3*40.6/13 (~= 9.37 A). I convolved 8A map with 10A
Gaussian for simpler presentation.
Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH.
Microtubule structure at 8 A resolution.
Structure. 2002 Oct;10(10):1317-28.
Tubulin atomic model.
Atomic model of alpha and beta tubulin
(PDB 1jff)
at 3.5A obtained by
electron crystallography of 2-D zinc-induced tubulin sheets. One GTP,
one GDP, and one taxol ligand. Protofilaments are anti-parallel one another,
unlike microtubule organization.
Lowe J, Li H, Downing KH, Nogales E.
Refined structure of alpha beta-tubulin at 3.5 A resolution.
J Mol Biol. 2001 Nov 9;313(5):1045-57.