How to Use Chimera: Ebola Virus
Tom Goddard
September 25, 2014
Introduction to some Chimera visualization and analysis capabilities for proteins and 3d electron microscopy,
looking at Ebola virus.
Prepared for Wah Chiu's class at Baylor.
Computational Mathematics for Biomedical Scientists:
"This course introduces essential computational and mathematical
concepts to students who are interested in computational biology and
bioinformatics. It is intended that each of the concepts will be
taught in the context of the real biological problems ranging from
genomics to structural biophysics."
BBC image of Ebola virus.
It doesn't really glow, and journal literature says it is usually about 10x longer than diameter,
about 80 nm by 800 nm.
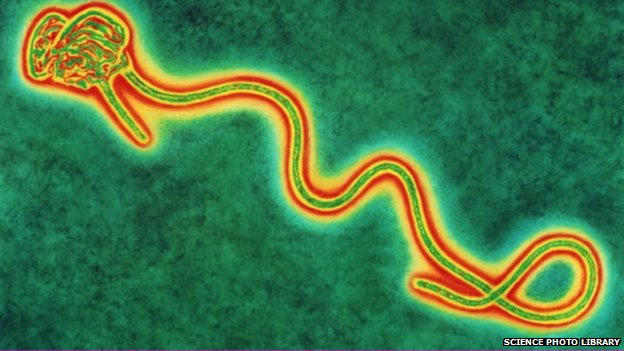
A more representative Ebola electron microscope image
from 1977 Ebola outbreak from W Slenczka.
Ebola virus illustrated by Ivan Konstantinov for 2011 Science magazine visualization challenge.
Image.

David Goodsell illustration of Ebola virus proteins. Protein Databank Molecule of the Month for October.
[Image removed pending publication.]

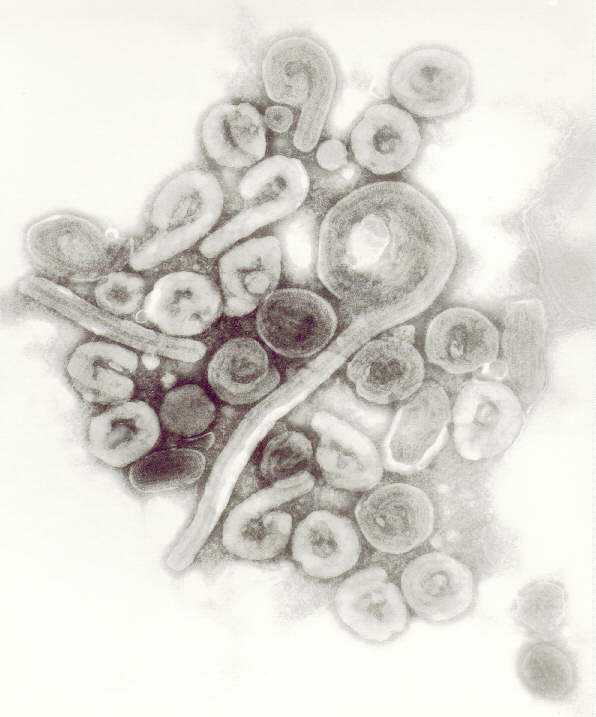
Electron microscopy of virus-like particles showing Ebola spikes, from
Spatial Localization of the Ebola Virus Glycoprotein Mucin-Like Domain Determined by Cryo-Electron Tomography,
July 2014.

Chimera demonstration looking at Ebola glycoprotein data

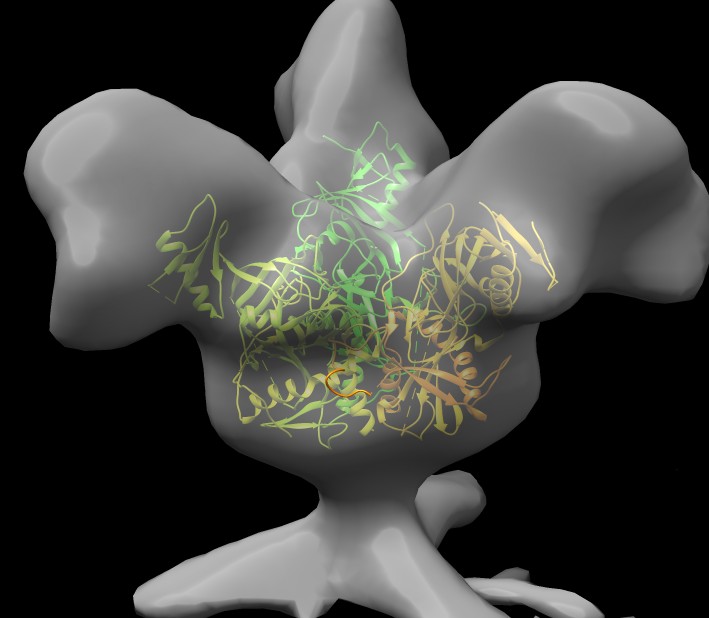
| 
| 
| 
|
|
Glycoprotein from subtomogram imaging from Sriram Subramaniam at NIH, July 2014.
EMDB 6003.
|
Find ebola glycoprotein on pdb web site, 3csy, open in Chimera, hide atoms, delete extra 1/3 of a trimer,
hide antibodies (~ribbon :.A-H), fit into EM map 6003 (need to select just glycrotein, not antibodies, drag select).
|
Antibodies bind near base of spike in 3csy.
|
Mucin-like domain was deleted from 3csy xray structure to aid crystallization, ~190 residues deleted
from looking at 3csy web page
sequence diagram and mousing over bars for glycoprotein chains.
Load EM map with mucin-like domain deleted,
EMDB 6004.
|
Recording a Movie
How to make an animation spinning the atomic model and map 360 degrees.
Use command to turn in 1 degree steps for 360 total steps.
turn z 1 360
This rotated about screen z axis. To use the density map z axis
turn z 1 360 coordinateSystem #0
Note that if we use the PDB coordinate
system instead (#1) it rotates about some weird axis from crystallographic coordinates.
Now add movie recording commands.
movie record ; turn z 1 360 coord #0 ; wait ; movie encode ~/Desktop/3csy_spin.mp4
Spin movie.
Chimera movie examples web page shows useful commands for movie making.
Find other glycoprotein structures
Are there other Ebolo glycoprotein structures that have the mucin-like domain?
Use Chimera BLAST search of PDB to find similar sequences, chain I is glycoprotein.

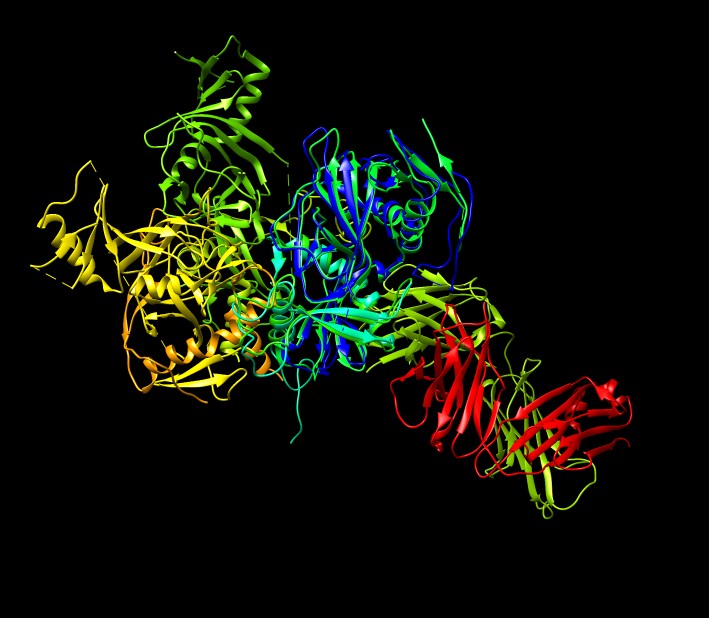
| 
| 
| 
|
|
BLAST shows two similar PDB structures
3s88
and
3ve0.
Select 3ve0 line and load with BLAST dialog Load Structure button so it is aligned with 3csy.
Color with "rainbow chain" command.
|
Hide 3csy shows we only have one glycoprotein not the 3 that make a spike.
PDB file only contains the unique part under symmetry. Show crystal unit cell.
Color all extra copies gray ("color gray #4-26") and show outline.
|
Unit cell is 195 Angstroms on a side according to dialog, approx 20 nm, so 50,000 copies
of this brick along each axis make a millimeter size crystal for x-ray diffraction.
Mouse over to see #7 and #11 make trimer with original copy, "color orange #7,11".
|
The antibodies in 3csy and 3veo bind to nearby locations ot the base of the spike. Why there?
Look at residue conservation.
|
Sequence conservation and antibody binding sites
Load sequences of several Ebola strains from past epidemics and color by conservation
(Multalign Viewer menu Structure / Render by Conservation). Select by hand blue strand
that is near 3csy antibody loops. Low conservation indicates this antibody KZ52 won't work
on the other strains besides the Zaire strain in 3csy, which is observed experimentally.
This antibody was obtained from a survivor of the 1995 epidemic. Vaccine to elicit this
antibody would not be effective across virus strains.


Would want an antibody that targets for example the nearby all red helix, conserved across strains.
ZMapp drug used experimentally on a few Ebola victims is a mix of 3 antibodies (c13C6+c2G4+c4G7)
against Ebola glycoprotein.















