Software for Interactive Analysis of Large Molecular Assemblies
Tom Goddard
November 13, 2006
Outline
- This core project continues one in our current NCRR grant titled
"Volume visualization for analyzing cellular systems at multiple
resolutions".
-
Goddard TD, Huang CC, Ferrin TE.
Visualizing density maps with UCSF Chimera.
J Struct Biol. 2006 Jul 15; [Epub ahead of print]
-
Goddard TD, Huang CC, Ferrin TE.
Software extensions to UCSF chimera for interactive visualization of large molecular assemblies.
Structure. 2005 Mar;13(3):473-82.
- Three Aims of this Presentation:
Describe 5-year vision and how far along we are in achieving
that vision.
See Chimera in action. Demonstration of capabilities for
interactive analysis of large atomic models and electron microscopy
density maps.
Progress since grant submission. Additional slides describe
progress since June 2006 emphasizing collaborative work. These will
not be covered in talk. Project direction has not changed.
Illustrating the Problem
From Tim Baker talk, Large Biomolecular Complexes Visualization
workshop, September 2005. Color PBCV-1 virus map to illustrate
symmetry.
Ten computer programs needed
-- several in-house utilities (RobEM, pif*), Situs, O, and Chimera.
"I do not know if I make myself clear, probably not. It is really a kind of
complex procedure."

Solution and Vision
- After workshop we began a collaboration with Baker lab.
- With Chimera additions in the last year this can be done entirely
in Chimera taking half an hour instead of half a day.
- Goal of our project is to make common visualization and analysis
tasks possible entirely within Chimera.
- This will enable studies of large assemblies by labs that do not have
their own programmers writing in-house software.
- The main impact will be that more labs can pursue studies of large
assemblies.
Project Vision
Our goal is to expand Chimera capabilities to encompass the common
interactive visualization and analysis tasks in the study of large
molecular assemblies involving atomic models and electron microscopy maps.
We will continue our leading and highly unique role developing
interactive software to supplant the patchwork of programs
borrowed from other disciplines in current use for exploring large
assemblies.
Keywords:
System integration.
Highly collaborative work.
Same-day prototyping.
Multiple length scales.
Real-time data exploration.
User interface.
Documentation.
Thousands of users.
Desktop computing.
Not the Vision:
Long-running computations.
Special-purpose algorithmic code.
Software for specialized hardware (computing clusters, volumetric rendering boards, ...).
Current Chimera Standing
- Approximately 500-1000 researchers currently use Chimera to study
large assemblies, primarily by electron microscopy. Estimate
is rough -- based on email and workshop contacts with ~100 users per
year combined with Chimera registration statistics (~2000 voluntarily
registered users, likely underestimates total users by factor of 2).
- In 2006, three journal covers for Science and Nature depicted viruses,
all three groups use Chimera for interactive analysis. We interact with
all three groups, two being collaborators
(Gabe Landers with Bridget Carragher, and Wen Jiang formerly with Wah Chiu,
Jonathan Hilmer in Brian Bothner lab).
EM Databases
The two public EM databases use Chimera as their
primary visualization tool, and link to Chimera web site on each of their
data set web pages.
- The two electron microscopy public databases are new and growing rapidly.
- EM DataBank (EBI) founded in 2002.
Number of entries doubled in past year (currently 268).
- Virus Particle Explorer (VIPERdb) EM archive founded in 2004.
- Almost all database images created using Chimera.
Gallery of EMDB entries
September 2005.
Example VIPERdb web page

Chimera Extensions from Chiu Lab
Labs can develop Chimera plugins to meet specialized needs, building on
existing Chimera capabilities.
Example: 25 Chimera plugins developed
by the National Center for Macromolecular Imaging and distributed with
EMAN. User interfaces shown below.
- Fitting (5 tools)
- Secondary structure search (4)
- Segmentation (5)
- Map filtering (2)
|
- Vibrational mode viewer
- Animation (2)
- EMAN interface
|
- Measuring Stick
- Surface coloring
- File conversion (3)
|
Other Software
Chimera is the only widely distributed interactive analysis program
focused on large assemblies and electron microscopy.
Each lab uses many software packages because none encompass
basic EM map and model capabilities.
Volume visualization packages, commercial, targeting diverse fields
medical imaging, earth sciences, oil/gas resevoirs, aerospace
engineering, financial modeling, .... Have very limited molecular model
capabilities.
- Amira - Focus on medical imaging.
Excellent density map capabilities. Limited molecular model features
documented in 35 pages versus ~500 for Chimera.
- IRIS Explorer -
Molecular model capabilities developed by Chiu lab (Baylor) have been
abandoned.
- AVS - Molecular model capabilities
developed by Olson lab (Scripps) have been abandoned.
- Vis5D -
climate modeling (non-commercial). Demonstrated by Steve Ludtke five
years ago for EM maps. Code recompiled for cube shaped EM maps instead
of pancake shaped atmosphere maps.
Molecular modeling packages. Have limited density map capabilities
or specialized for atomic resolution crystallographic maps.
- O.
Standard package for interactive model building in crystallographic
density maps.
- PyMol.
Drug binding, pharmaceutical industry focus.
In-house EM map packages. Inadequate EM lab resources to develop and
distribute program for wide base of users.
- RobEM.
Michael Rossmann (Purdue), Tim Baker (UCSD, formerly Purdue).
- Priism.
Dave Agaard, John Sedat (UCSF).
Summary of Proposed Developments
Require a range of map and model analysis capabilities focused on
the organization of molecular assemblies beyond the atomic level:
arrangements of molecules, domains, and secondary structure elements,
their mechanisms of assembly and conformational rearrangements.
Large Atomic Models
- Library of standard symmetries.
- Buried surface area calculations for unique molecular interfaces.
- "Explode" subassemblies to examine interfaces.
- Finer and coarser grouping for surfaces.
- User interface improvements for exploring quaternary structure.
- User-defined meaningful subassemblies.
Animating Assemblies
- Commands to create many types of smooth transitions: motion interpolation,
fade in/out, color transitions, style transitions, motion with clash avoidance
for any atoms, residues, domains, molecules, or subassemblies.
- Graphical time-line movie composer.
- Canned one-button movies: spin 360 degrees, move clip plane through map.
Single Particle EM Maps
- Steric clash avoidance in fitting atomic models.
- Fitting individual secondary structure elements and domains with
energy minimization (core 1) to repair connections.
- Interactive watershed type segmentation.
- Correlation measures between maps or map subregions and fitting maps into other maps.
- Masking with arbitrary surfaces, e.g. icosahedral shells.
- Optimizations for large maps (100 Mbytes - several Gbytes) on desktop computers.
Structurally Heterogeneous Assemblies (primarily EM Tomography)
- Map filtering and denoising (median filter, bilateral filter, inversion).
- Segmentation of filaments using tubular paths.
- Measurements of angles, symmetry (e.g. helical pitch), masses.
- Volume time-series for in vivo light microscopy. Interactive and
semi-automated feature tracking.
Chimera Demonstrations
- Large atomic-resolution model. Examine Human Rhinovirus 2 contacts
with a fragment of its cellular receptor.
On web.
- Single particle electron microscopy map. Fit a crystal
structure in a myosin thick filament map.
- Electron microscope tomography. Segment the conical HIV1
core particle from enveloped virus.
HIV Spike EM Tomography
June 2006 EM tomography example fitting HIV gp120/gp41 spikes using Chimera.
Spikes recognize target cells and initiate viral fusion.
Illustrates combined use of tomography and structure averaging approach.
Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH.
Distribution and three-dimensional structure of AIDS virus envelope spikes.
Nature. 2006 Jun 15;441(7095):847-52. Epub 2006 May 24.
John Sedat Collaboration, EM Tomography
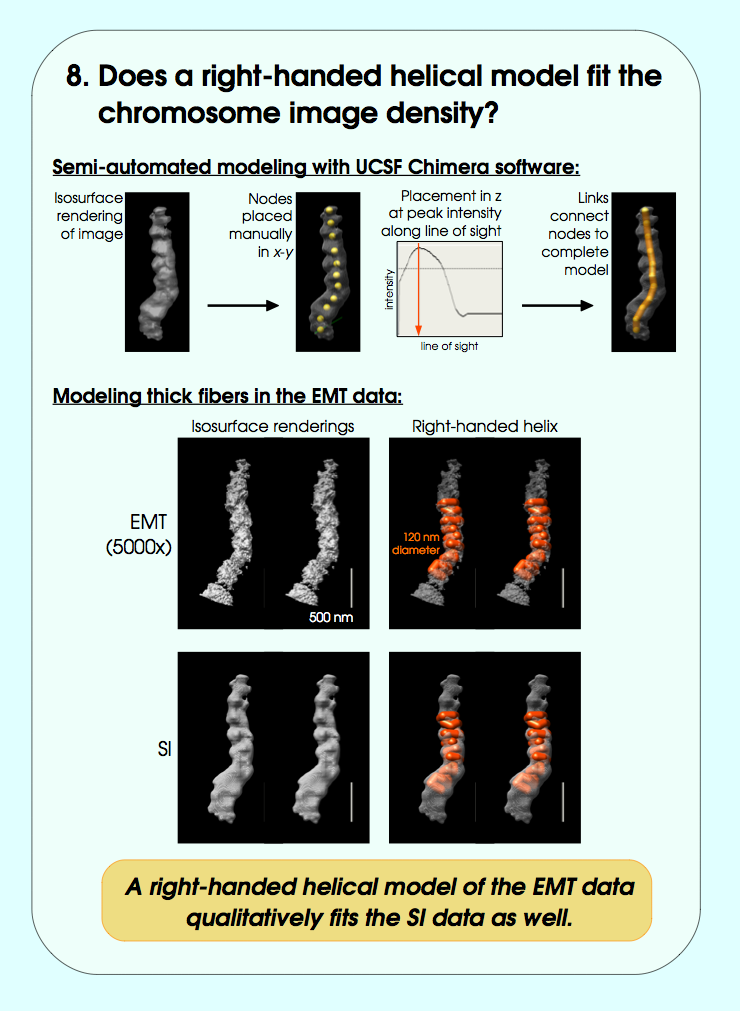
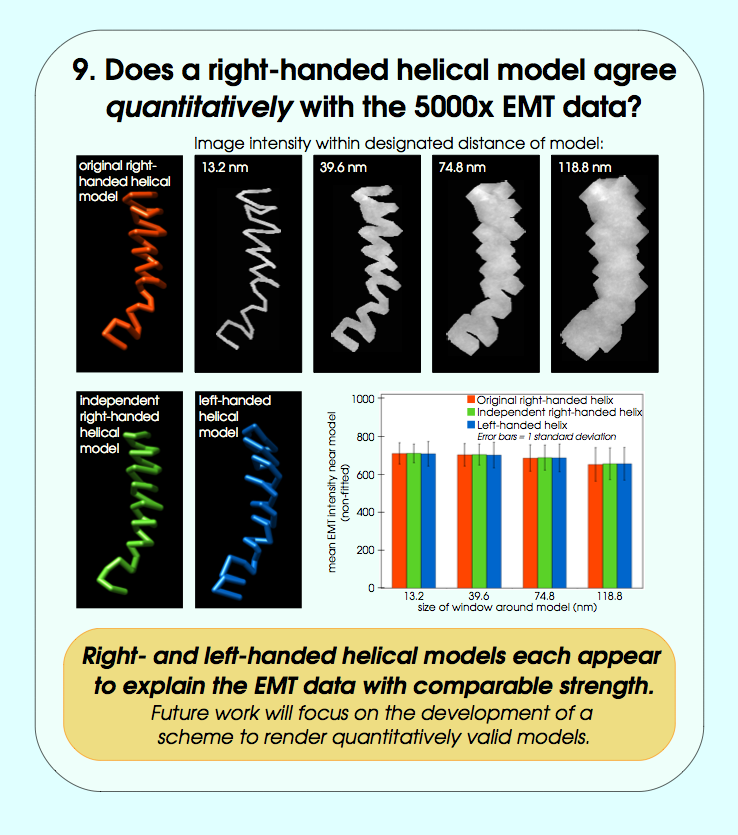
September 2006 chromosome EM tomography poster from Sedat lab.
Chimera used for modeling helical structure.

Manfred Auer Collaboration, EM Tomography
Recent poster showing EM tomography of haircell molecular mechanical linkers.
Chimera used to model unknown linkers and fit candidate molecules.


Progress since June 2006
Example Chimera scripts and commands developed for collaborators since
June 2006 grant submission. These are plugins distributed separately
from Chimera on our
experimental features
web page.
- Scripts to auto-generate virus EM map images and movies, Padmaja Natarajan, Jack Johnson lab, Virus Particle Explorer Database (VIPERdb).
| Radially colored.
| Cut in half.
| Movie showing slab at progressive depths.
|

| 
| 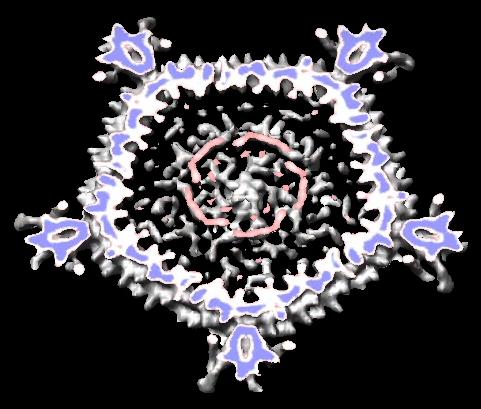
|
- Create icosahedral schematic cages for virus capsids, Jinghua Tang, Tim Baker lab, also VIPERdb request.
- Display clashes between asymmetric units in crystals and biological assemblies, Cathy Lawson, Protein Data Bank (PDB).
- Split density map into separate map for each color, Steve Ludtke, National Center for Macromolecular Imaging (NCMI), and used for John Sedat lab chromosome data.
- Neural dendrite spine length measurement, confocal microscopy,
IMOD segmentation file reader, Maryann Martone, National Center for Microscopy and Imaging Research (NCMIR).
Collaborations Initiated from Workshops and Conferences
Many collaborations outside UCSF have been established by attending
meetings and giving talks. Here are examples from the past year.
- Large assembly visualization workshop, UCSD, Sep 2005. Met Tim Baker. All experimentalists at meeting using Chimera.
- Automated Molecular Imaging Forum, Dec 2005. Spent 2 days with Baker lab initiating collaboration. Met Bridget Carragher, NRAMM director.
- Biophysical Society Meeting, Salt Lake, Feb 2006. Worked out database viewer collaboration with Bridget Carragher, NRAMM.
- BioImage Informatics, Santa Barbara, July 2006. Possible Maryann Martone, Mark Ellisman NCMIR collaboration, microscopy of dendrites.
- We host collaborator visits. VIPERdb visit from Padmaja Natarajan, Aug 2006. Day long discussion of possible new tools.
- CryoEM Standards Task Force, Fall 2006, teleconferences. Involves several existing collaborators: Henrick (EBI) / Berman (PDB) / Chiu (NCMI).
- Future: CryoET of Cells workshop, Max Planck Institute, Dec 2006, hosted by Wolfgang Baumeister. They wish to develop EM tomography extensions for Chimera.
Collaborations with Public Databases
Collaborations with public archives are important for success of
Chimera and the databases.
Omitted collaborations with EMDB and VIPERdb public electron
microscopy databases from grant proposal.
- Protein Data Bank. Cathy Lawson, Helen Berman (RCSB).
Remediation of biological unit and crystal unit cell meta-data.
- Electron Microscopy Databank. Richard Newman, Kim Henrick (EBI).
Density map formats, meta-data, and web images.
- Virus Particle Explorer Database.
Padmaja Natarajan, Jack Johnson, Vijay Reddy (MMTSB resource).
Automatic image generation for atomic models and EM maps.
- EMDB and VIPERdb database web pages link to Chimera web site.
Core Project Aims
Broad aims of large assemblies core project listed in our June 2006 grant proposal.
Develop advanced capabilities for:
- Display and manipulation of atomic models comprised of tens to thousands of macromolecules.
- Creating animations of assemblies such as virus capsids and ribosomes that elucidate structural organization and conformational changes.
- Display and analysis of medium and low resolution density maps intended primarily for researchers studying molecular assemblies using electron microscopy single particle reconstruction.
- Analysis of structurally heterogeneous assemblies, for example chromosomes or polymorphic viruses such as HIV, imaged by electron microscope tomography.