Molecular Representations
Several kinds of molecular representations are available in Chimera:
- wire - simple line drawing with dot atoms and wire bonds
- stick - "endcap" atoms and stick bonds
- ball-and-stick - ball (scaled
VDW radius) atoms and stick bonds
- sphere - sphere (full VDW radius)
atoms and wire bonds; also called Corey-Pauling-Koltun (CPK)
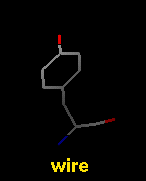
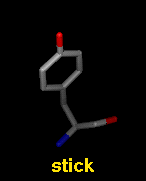
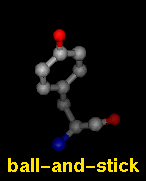
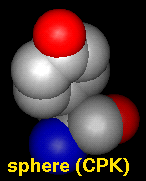
The draw mode is an attribute of individual
atoms and bonds, whereas wire linewidth is a
model attribute.
Thus, different parts of a single model cannot have different
linewidths, although they can be in different representations.
Proteins and nucleic acids can be drawn as a ribbon.
Protein secondary structure can also be shown with
PipesAndPlanks, and special representations of the
base and sugar components of nucleic acids (combined with
sticks in the figure) can be generated with the
Nucleotides tool.
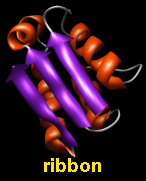
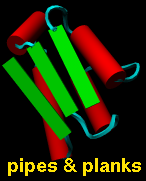
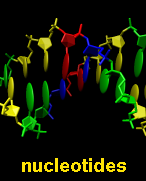
Ribbon display is an attribute at the residue level.
The pipes-and-planks and special nucleotide representations
are VRML objects and can only be displayed/undisplayed as a whole.
There are several ways to change or add representations:
- the Actions menu can be used to
switch among wire, stick, ball-and-stick, and sphere representations
of atoms and bonds, change wire linewidth, add a ribbon to models
containing protein or nucleic acid, or change which style of ribbon is shown
- the command represent can
be used to switch among wire, stick, ball-and-stick, and sphere representations
of atoms and bonds
- the command linewidth controls
the thickness of lines drawn to show bonds in the wire representation
- the command ribbon can be used to
add a ribbon to models containing protein or nucleic acid
- the command ribrepr can be used to
change which style of ribbon is shown
-
Ribbon Style Editor can be used to edit ribbon
scaling (width/height),
style (the general shape of the cross-section), and
residue
class (which atoms guide the ribbon path);
scalings, styles, and residue classes can be named and saved
- the molecule model attributes panel
can be used to:
- switch among wire, stick, ball-and-stick, and sphere representations
of atoms and bonds
- change wire linewidth and line type (can even be dotted or dashed)
- change stick and ball scale factors
- add a ribbon to models containing protein or nucleic acid
- change which style of ribbon is shown
- the pseudobond attributes panel can be used to:
- switch between wire and stick representations of
pseudobonds
- change wire linewidth and line type
- change stick scale factor
- after a selection of atoms, bonds, and/or
pseudobonds is made, its representation can be altered with the
Selection Inspector
-
PipesAndPlanks can be used to add a VRML pipes-and-planks
representation to models containing protein
-
Nucleotides can be used to add VRML representations
of the sugar and base components of nucleic acids
In Chimera, all of the representation types
can be manipulated in real time, unlike the space-filling models
created with the commands conic,
neon, and
ribbonjr.
Molecular Surfaces
Molecular surfaces can be created, displayed, and manipulated in real time.
A molecular surface can be generated with the Surface section of the
Actions menu or
the command surface.

The molecular surface representation can be switched among
solid, mesh, and dot using:
Molecular surfaces produced by Chimera are created with
embedded software from the
MSMS package by
Prof. Michel Sanner, described in:
M.F. Sanner, A.J. Olson, and J.C. Spehner,
"Reduced surface: an efficient way to compute molecular surfaces"
Biopolymers 38:305 (1996).
A molecular surface consists of probe contact
plus reentrant surface; this is different from the surface traced out
by the probe center (often called the solvent-accessible surface).
The probe radius and density of vertices can be adjusted in the
surface attributes panel.
Dot molecular surfaces in MS/DMS format
(calculated and written out by a separate program)
can also be displayed in Chimera.
Other Surfaces
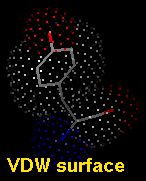
A van der Waals (VDW) surface differs from a molecular surface
in that fine crevices are not smoothed. A VDW dot surface can be
displayed with the command vdw
and its dot density adjusted with
vdwdensity. The VDW dot size and dot density can also
be adjusted in the the
molecule model attributes panel.
Note that the sphere representation
of atoms also reveals the VDW surface.
Other types of surfaces that can be shown in Chimera are: