Unit Cell 
Unit Cell builds crystallographic unit cells
using information in PDB files.
There are several ways to start
Unit Cell, a tool in the Utilities category.
A crystallographic unit cell consists of a unique set of coordinates,
duplicated and transformed according to the crystallographic and
(if present) noncrystallographic symmetries in the crystal.
Unit Cell can be used to regenerate the full unit cell, or only
those parts defined by
crystallographic symmetry or noncrystallographic symmetry.
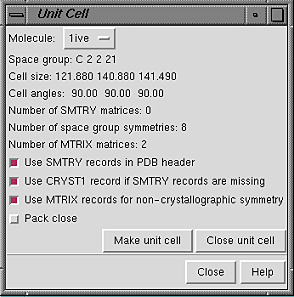 Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.
Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.
Make unit cell loads copies of the coordinates from the
same source as the originally opened model (a local file or a file
fetched from the web) and transforms them using the symmetry information
specified by the checkbox settings.
Pressing the button a second time will
create a new set of copies without deleting the first set.
Close unit cell deletes all
open copies of the molecule, except for the original.
All copies will be deleted, not just the ones
specified by the current checkbox settings.
The checkbox settings control which matrix
information is used to determine the number of copies and
their transformations:
- Use SMTRY records in PDB header
- The SMTRY1, SMTRY2, and SMTRY3 records found in REMARK 290
contain the rows of transformation matrices defining the
crystallographic symmetry.
These records are not present in all PDB entries.
- Use CRYST1 record if SMTRY records are missing
- The CRYST1 record contains the unit cell size in angstroms, the cell angles,
and the name of the space group. The transformations defining the
crystallographic symmetry can be looked up in a table by space
group name. Some PDB entries contain nonstandard space group names
that are not in the table. When this occurs,
the number of space group symmetries is reported as zero; if the
SMTRY matrices are also missing, then the crystal packing is unknown.
- Use MTRIX records for non-crystallographic symmetry
- The MTRIX1, MTRIX2, and MTRIX3 records contain the rows of transformation
matrices defining the noncrystallographic symmetry.
Only a small fraction of
PDB entries have MTRIX records (~10% in 2003). Most of these describe how
to transform the coordinates of one chain to match closely the coordinates
of another chain already present in the PDB file. In a few cases (~0.25%
of PDB entries), the MTRIX records describe a transformation that produces
a new copy whose coordinates are not already in the file.
These two cases are distinguished by the contents of
column 60 in the MTRIX records. A "1" means that
the transformation yields coordinates that are already given (these
MTRIX records are ignored by Unit Cell), whereas a blank space
means that the transformation will produce a new copy.
MTRIX column 60 is set incorrectly in some files; for example,
it is "1" in 2btv even though the MTRIX records
define coordinates not present in the file. Further,
MTRIX records are sometimes missing. For example, 1cd3 has no MTRIX
records, but the remarks describe how to produce them from the BIOMT
records. Of course, this cannot be handled by Unit Cell.
MTRIX records do not describe crystallographic symmetries, but
define additional symmetries of the asymmetric unit of the crystal.
Because the copies of the molecule occupy nonequivalent positions
in the crystal, they usually have small structural differences due
to differing crystal contacts.
Thus, an independent set of coordinates is usually included for these copies,
and MTRIX records are not needed.
MTRIX records are often used for icosahedral virus particles, where
the PDB file will include the coordinates of one molecule in the shell
and MTRIX records describing how to place copies that are
not crystallographically equivalent.
The size of the virus particle precludes independent refinement
of coordinates for each copy of a molecule in the shell.
- Pack close
- The transformation matrices frequently specify copies that are
spread out in space rather than tightly packed. Any copy
can be shifted along a crystal axis by an integral multiple of the
cell size along that axis, which can result in a more tightly packed
(but equally valid) unit cell. Pack close indicates that
copies should be shifted to increase compactness the next time
Make unit cell is pressed. To perform close packing,
the center of the bounding box of the original molecule is calculated.
The center is copied to the new positions along with the atomic coordinates.
Each copy is then shifted so that its center is within half a unit cell
length of the center of the original molecule along each axis.
The unpacked positions are often symmetrically
located, while the packed positions may not be.
Limitations
As mentioned above, many problems are due to information that is missing
from or incorrect in the PDB file.
Unit Cell does not generate multimers defined by BIOMT records,
but this can be done with
Multiscale
Models.
There is little control over which part of the crystal packing is shown.
Given an X-ray density map, for example,
there is no way to make only the copies that intersect the map.
There is no way to force specific copies to be adjacent
so that their packing interface can be examined
(one copy may be one cell-width away from the desired position).
However,
Multiscale
Models can generate a 3 by 3 by 3 block of unit cells, which
may allow the interface of interest to be examined when the
two monomers are shown in atomic detail.
UCSF Computer Graphics Laboratory / July 2003
 Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.
Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.