Chimera Workshop
November 17-18, 2005

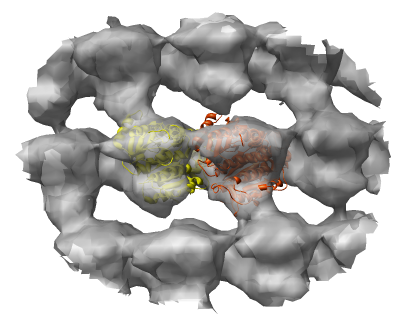
EMD 1130 is GDP bound bovine microtubule at 12 angstroms.
Image shows EMD 1129 (inner layer) and EMD 1130 (outer layer) of double walled microtubule. Yellow and orange balls are alpha and beta tubublin proteins.
1jff is a bovine alpha/beta tubulin by electron diffraction of 2D taxol stabilized sheets, 3.5 angstroms.
2bto is bacterial tubulin called btuba thought to be horizontally transferred from eukaryotes, found in only one bacteria, has 30-40% seq identity and RMSD 1-2 angstrom differences. Chose this one because crystallographic data (2.5 angstroms) available.
Beta tubulin is on the left, alpha tubulin on the right (1jff). Slightly different loop shapes.
Use surfcat A :.A and surf A to surface individual chain.


Use different colors for alpha and beta electrostatics to easily see the boundary between them.


The previous bovine tubulin structure was determined by electron diffraction of a 2-D crystal, for which I don't have a density map. Look at bacterial tubulin (2bto) with density map from EDS.
Color model orange-red for contrast with white density.
Restrict density map display with surface zone to chain A. Use invert selection and hide atoms to restrict model display.


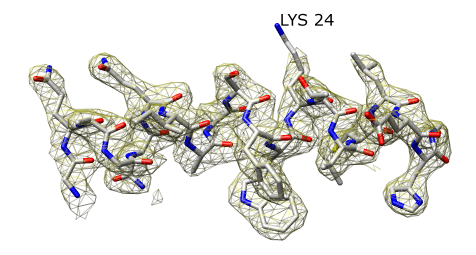
Use Sequence dialog (Tools -> Structure Analysis -> Sequence) to select first alpha helix. Restrict map and model to it. Make mesh transparent and yellow.
Show lysine sticking out. Hover mouse to identify residue.
Show alpha and beta monomers as ribbons and color yellow and orange.
Move into density map, select all atoms, zone at 50 A radius (for faster rotation), make map transparent, and optimize fit. Clear selection before inspecting fit (for faster rotation and clearer view).
Can try fit shifted right by 1 monomer. Slightly different fit. Would be interesting to try fitting each monomer separately.
The EMD 1130 map appears to have incorrect handedness compared to the Nogales paper images.